ML Build and Predict: Optimal Property-Predicting Graph Convolutional Neural Network Model
This tutorial guides you through building an optimal Graph Convolutional Neural Network (GCNN) model using the latest floe in the Machine Learning Model Building package. The GCNN floes are designed to streamline small molecule property prediction for drug discovery by simplifying the process of building and using deep learning models for molecular property prediction.
The GCNN model building floe offers several key features:
Comprehensive Featurization: Uses all relevant OEChem features for detailed molecular insights.
Automated Architecture: Automatically sets the optimal GCNN architecture based on your input data, saving you time and effort.
Actionable Reports: Provides a statistical Floe Report with guidance to prevent overfitting and easily train models.
Optimized Performance: Automatically chooses effective convolution layers and batch size for your specific task.
Scalable: Leverages PyTorch DDP to efficiently train on large datasets across multiple GPUs.
This package helps chemists quickly explore chemical space, prioritize promising candidates, and gain deeper insights, thereby accelerating the drug discovery pipeline.
This tutorial uses the following floes:
Floe Input
This floe uses the PyrrolamidesA dataset for P1. This input dataset contains several OERecords. The floe expects two things from each record:
An OEMol, which is the molecule to train the models on.
One of the following:
A Float value, which contains the regression property to be learned. For this example, it is the NegIC50 concentration.
A Int value, which contains the classification int property to be learned. For this example, it is the IC Class.
A Str value, which contains the classification string property to be learned. For this example, it is the IC Class.
Here is a sample record from the dataset:
OERecord (
*Molecule(Chem.Mol)* : c1cc(c(nc1)N2CCC(CC2)NC(=O)c3cc(c([nH]3)Cl)Cl)[N+](=O)[O-]
*NegIC50(Float)* : -2.6
*IC Class(Int)* : 1
*IC Class(Str)* : 1
)
Input Data
Preparing Your Data for GCNN Training
Choose the Data Processing of Small Molecule for ML Model Building Floe. Click “Launch Floe” to bring up the Job Form. The parameters can be specified as below.
Input Small Molecules to Train Machine Learning Models On: Select the PyrrolamidesA dataset or your own dataset.
Outputs: The output will be saved to the names in these fields, so change them to something meaningful to you. We are interested in Output Collection Name for Graph Feature Vector for our next step, as it will serve as the input for GCNN floe. We will use the defaults for this tutorial. This is essentially the “featurization” step that converts your molecules into a format that the GCNN can understand.
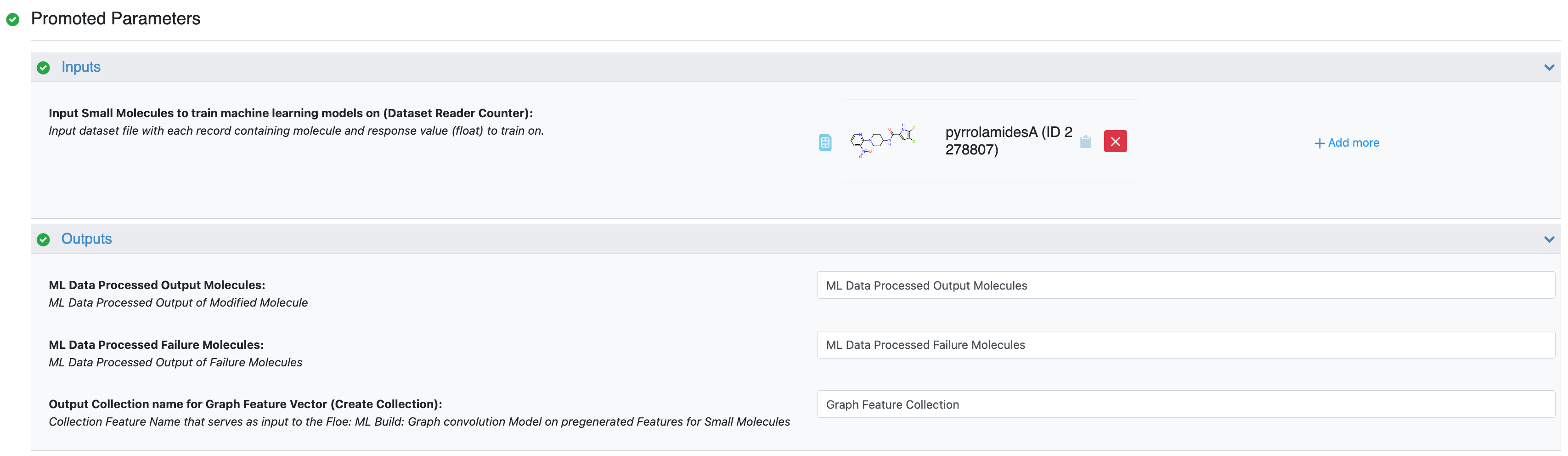
Figure 1. Input and output parameters for the Data Processing of Small Molecule for ML Model Building Floe.
Response Value Field: This in the property to be trained. The drop-down field generates a list of options based on the input dataset. For our data, choose NegIC50 in one run (regression) and IC Class for another separate run (classification).
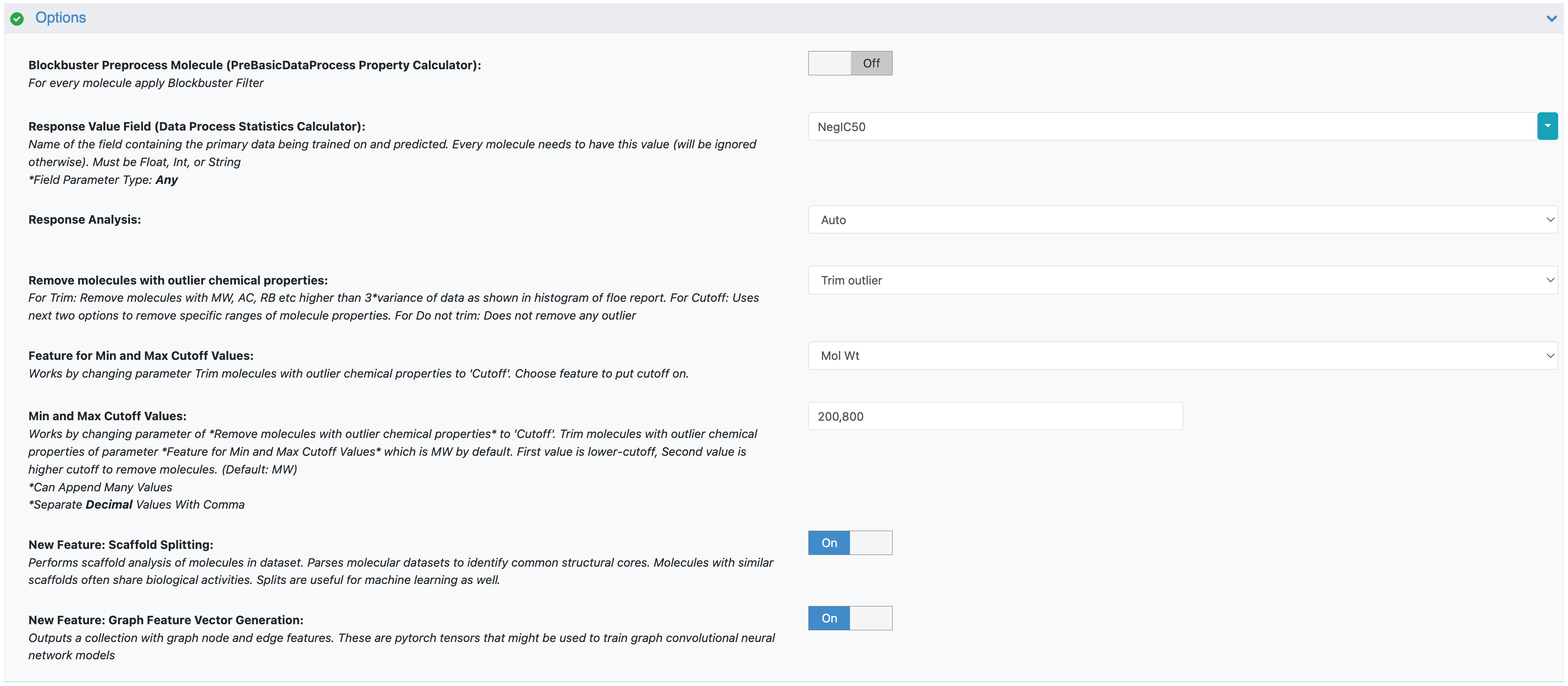
Figure 2. Additional parameters for the Data Processing of Small Molecule for ML Model Building Floe.
That’s it! Let’s run the floe. Click the “Start Job” button.
Connecting Data to Your GCNN Builder
Choose the ML Build: Graph Convolution Model on Pregenerated Features for Small Molecules. Click “Launch Floe” to bring up the Job Form. The parameters can be specified as below.
For the Collection: Input Graph Featurized for Small Molecules to Train GCNN ML parameter, use the output collection from your previous run of the Data Processing of Small Molecule for ML Model Building Floe.
When running this GCNN Floe for the first time, it is recommended to stick with all the default settings and automations. A key automation is the Automatically Adjust Hidden Layer Size parameter. This setting helps determine appropriate sizes for the graph convolution and fully connected layers within your GCNN, dynamically adjusting them based on the size of your input data. Please refer to the How-to Guide for the ML Build: Graph Convolution Model on Pregenerated Features for Small Molecules Floe for details on how to tweak this parameter.
That’s it! Let’s run the floe. Click the “Start Job” button.
The output from the Data Processing of Small Molecules for ML Model Building Floe, specifically the collection generated under the Output Collection Name for ML Build: Graph Convolution parameter serves as the input for your training and validation data in this GCNN Floe. This separation of data generation from model building floes ensures modularity, allowing you to reuse your preprocessed molecular data collection multiple times while iterating and optimizing your GCNN model’s architecture or training parameters. After the floe is completed, analyze the Floe Report (see the guide for How to Build Optimal Property-Predicting Graph Convolutional Neural Network Machine Learning Model by Tweaking Neural Network Architecture.
Floe Output
The floe outputs two data records.
Output Dataset: GCNN Built Model: This is the output GCNN that is to be used for prediction. It contains the field that includes metadata and the model tensor as shown in Figure 3.
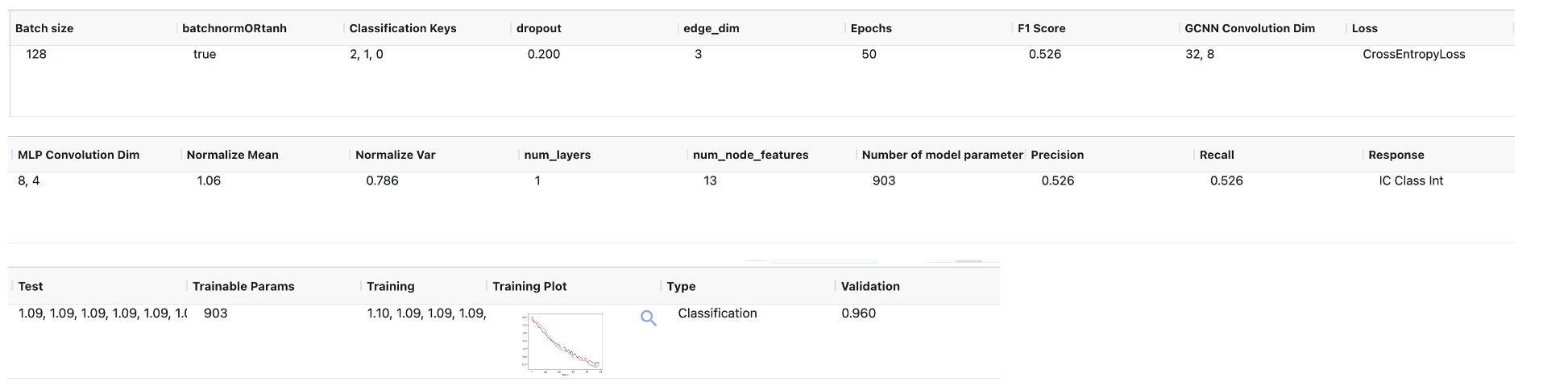
Figure 3. Output Dataset: GCNN Built Model.
Output Dataset: GCNN Validation Data: This is the output validation on scaffold split molecules. It outputs a prediction value if scaffold split is chosen. (With more parameters, you can choose the kind of split used.) It also adds the “Actual” and “Validation” fields to the input records.
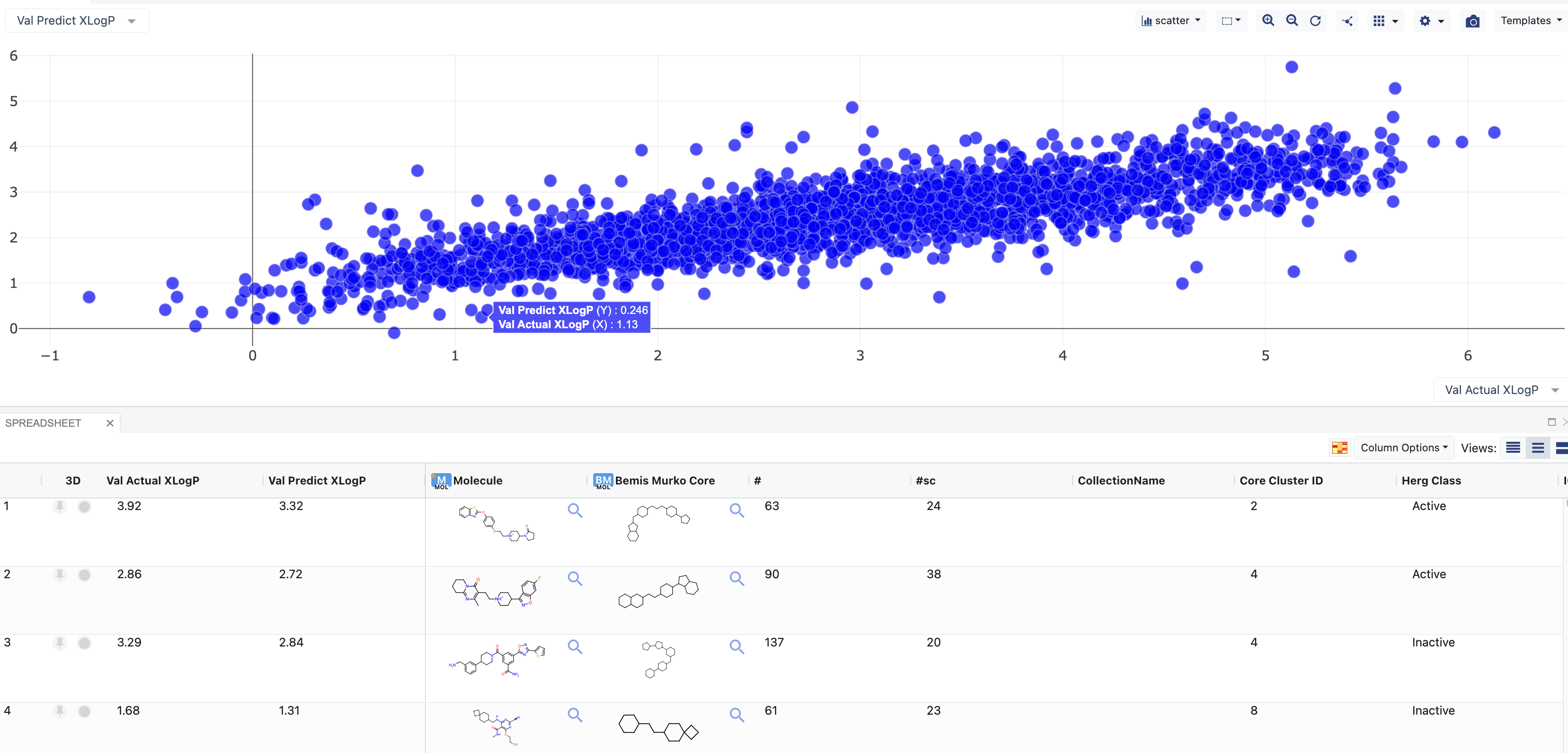
Figure 4. Output Dataset: GCNN Validation Data.
ML Predict
Choose the ML Predict: Graph Convolution Model Prediction Floe. Click “Launch Floe” to bring up the Job Form. The parameters can be specified as below.
Choose Input Molecule Dataset, which should be a molecule dataset. We can utilize the pyrrolamide dataset which we used above for training.
Input Data
For the Input File for Graph Convolution Model, use the GCNN Built Model from the previous run of the ML Build: Graph Convolution Model on Pregenerated Features for Small Molecules Floe.
Response Value Field: This field validates a prediction with actual values if that is included in your dataset. The drop-down field generates a list of options based on the input dataset. Choose from NegIC50 or IC Class based on what you chose above.
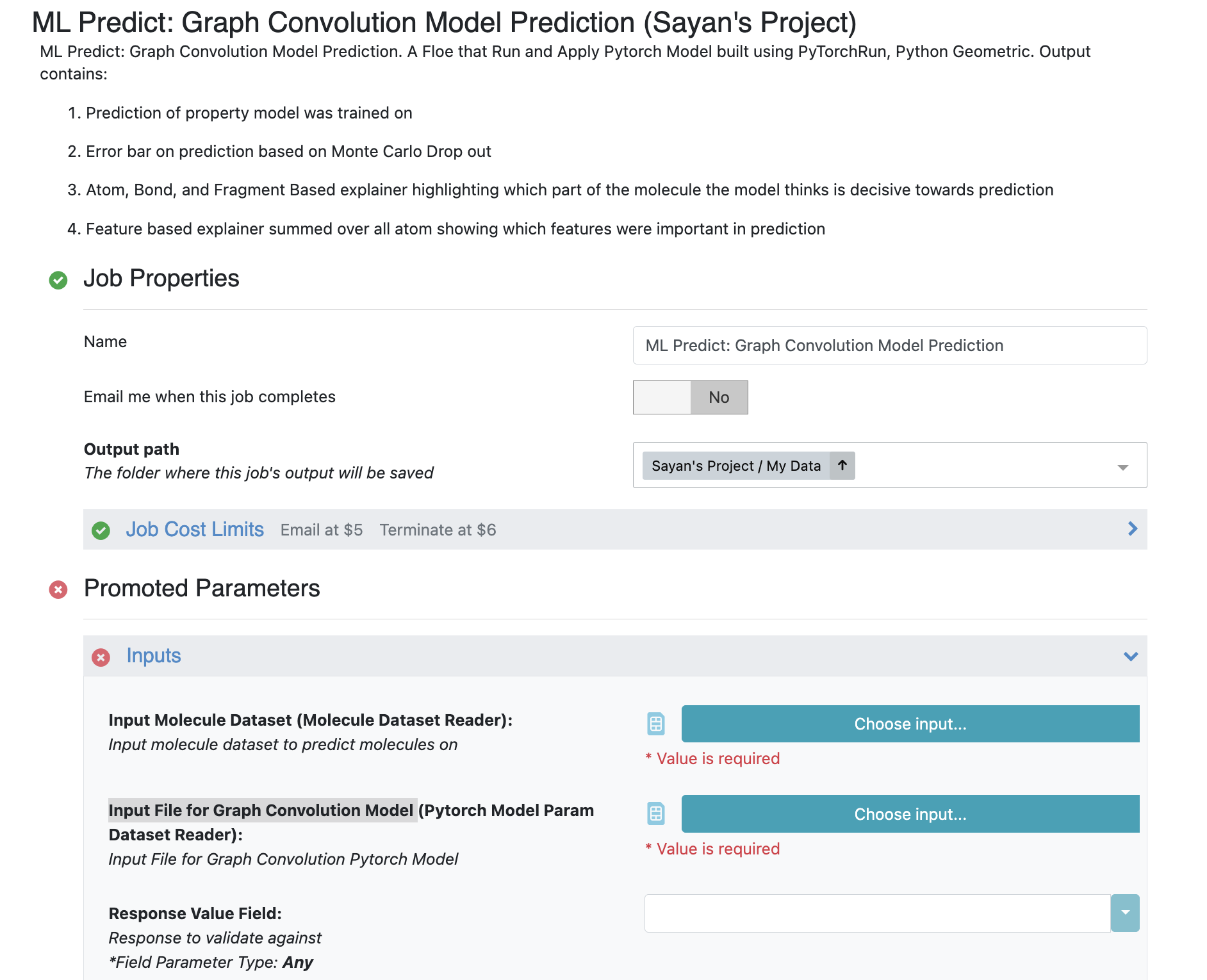
Figure 5. Floe options in the Job Form for the ML Predict: Graph Convolution Model Prediction Floe.
That’s it! Let’s run the floe. Click the “Start Job” button.
Analyzing the Output
Activate the output dataset so it can be viewed in the 3D & Analyze page.
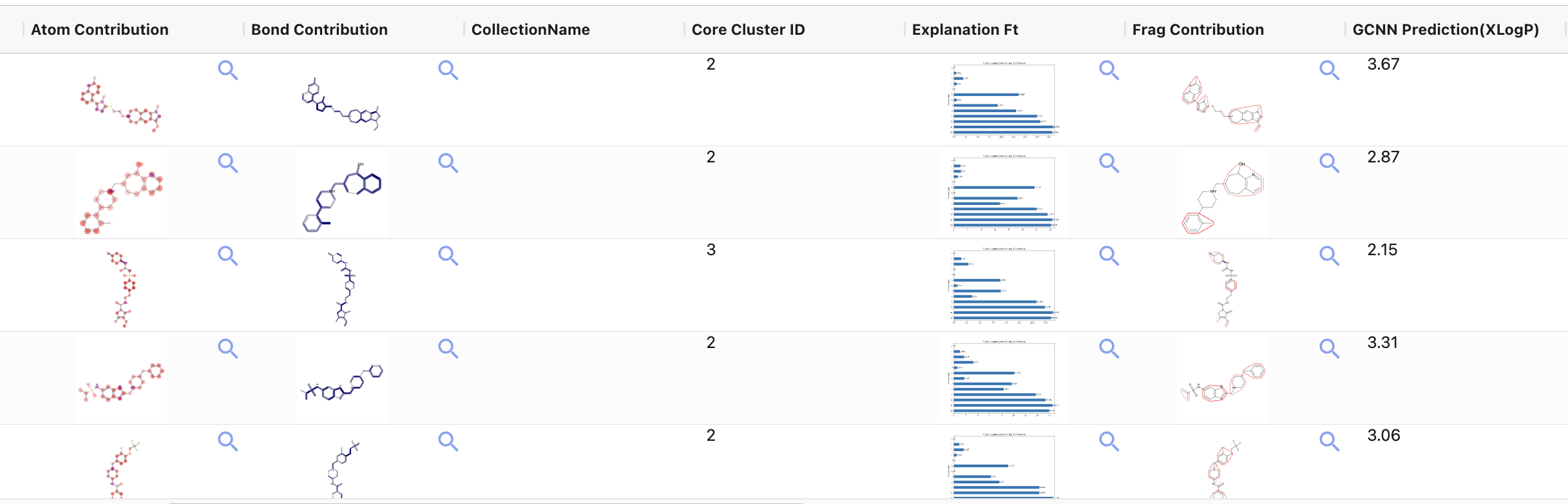
Figure 6. The GCNN predictor and explainers.
It contains the following fields:
GCNN Prediction (response): This is a prediction with a response name.
Error bars: This is attached as a metadata file to each prediction and can be seen in the plot.
Explainable AI (XAI)
Atom Contribution: This indicates which atoms contribute more strongly towards prediction.
Bond Contribution: This indicates which bonds contribute more strongly towards prediction.
Feature Vector Importance: The feature (bond or atom) the model was trained on has a greater contribution to the prediction. Note that the name order is same across all rows for easy comparison.