Bespoke and Custom Force Field
In molecular simulations, a bespoke force field is a set of parameters specific to a particular ligand, such as: bonds, angles, torsion angles and other parameters. It takes precedence over corresponding parameters in a general force field file for that particular ligand. In contrast, a custom force field is a comprehensive force field (typically a general force field with additional customized parameters) that will be applied to the entire ligand dataset. Incorporating bespoke force field parameters can improve the accuracy of molecular simulations, especially for molecules that are not well-described by general force fields. To associate bespoke parameters with the ligand dataset, run the Associate Force Field Offxml Files To Ligand Record Floe. The output dataset with bespoke parameters and/or a set of custom force field files can be applied in any of our MD floes, such as: Nonequilibrium Switching, Short Trajectory Molecular Dynamics, and PlainMD.
Note
The purpose of this tutorial is to apply bespoke and/or custom force field, rather than generate one.
Example Case: Biphenyl and ring substituents
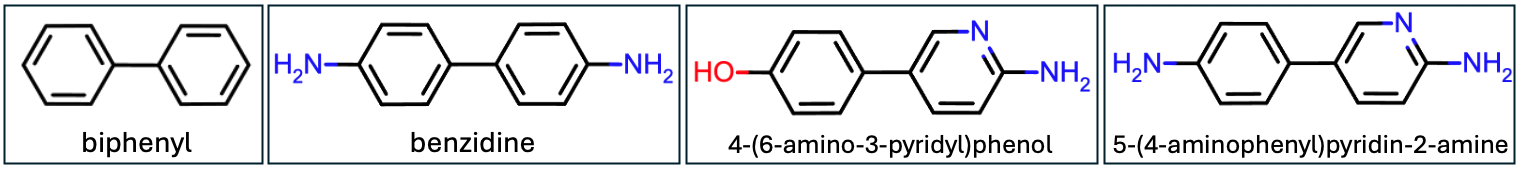
Figure 1: The chemical structures of biphenyl and its ring substituents
An example case that demonstrates the importance of customized force field parameters is the biphenyl molecule. When various substituents are added at the meta, ortho, or para positions, it impacts the conjugation of the molecule which has a direct effect on the energetic minima of torsion angles. While general force fields might be suitable for many scenarios, they might fall short in accurately representing conjugated systems. This limitation becomes apparent when running a basic energy minimization using a general force field. The results, as presented in Table 1 and Figure 2, reveal that the molecular mechanics minimized energies for all four variants are nearly identical. This similarity arises because the exact same torsion parameters are applied, neglecting the nuanced electronic properties introduced by distal substituents. By applying bespoke force field parameters that account for the specific electronic characteristics of each substituent, the dihedral angles between the two phenyl rings are distinct, showcasing the importance of bespoke force field parameters. In this tutorial, we won’t go into the specifics of creating BESPOKE parameters, but you can find more information here: OpenFF BespokeFit
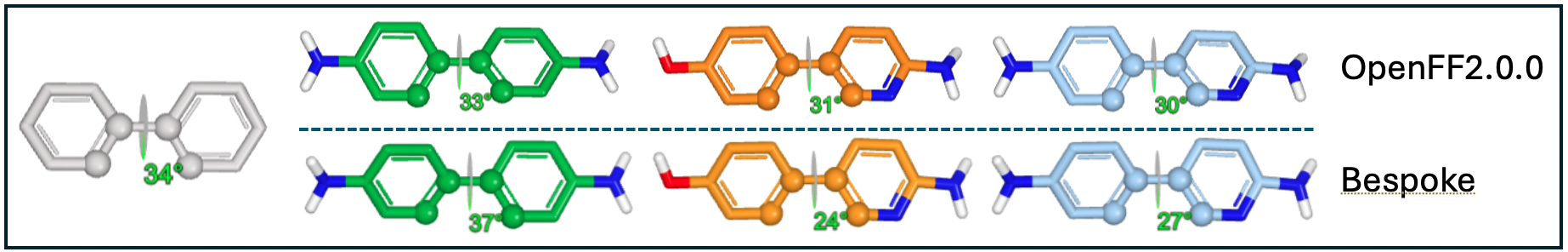
Figure 2: Biphenyl and its ring substitutents dihedral angles when using different force field
Table 1: Biphenyl dihedral angles
Ligand Name |
biphenyl |
benzidine |
4-(6-amino-3-pyridyl)phenol |
5-(4-aminophenyl)pyridin-2-amine |
|---|---|---|---|---|
Approximate Expected Torsion Angle [*] |
33 |
38 |
planar |
planar |
Minimization using OpenFF2.0.0 |
34 |
33 |
30 |
31 |
Minimization using Bespoke |
34 |
37 |
24 |
27 |
Once the bespoke parameter files are created, upload the bespoke offxml files and ensure you have a 3D structure of your ligand/molecule. Go to the Associate Force Field Offxml Files To Ligand Record Floe and use the ligand dataset and the bespoke offxml files as inputs to affiliate the bespoke parameters with your ligand dataset. An additional example case for the use of bespoke parameters can be found in this paper [Horton2022].
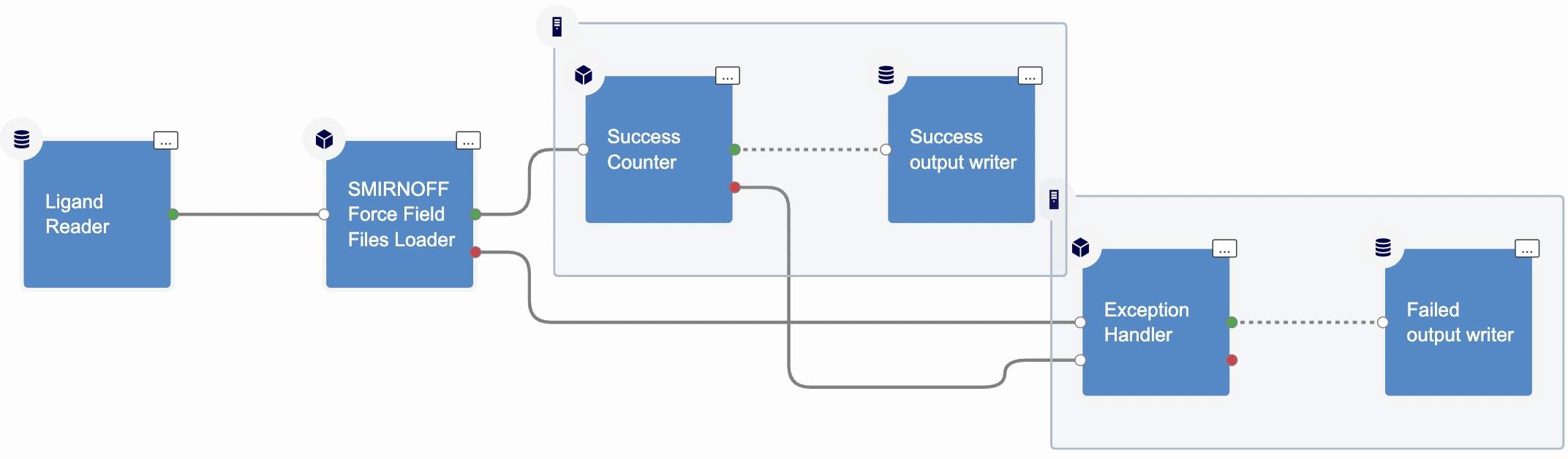
Figure 3: Associate force field offxml files with ligand record floe
The Floe Inputs
Running the Associate Force Field Offxml Files To Ligand Record Floe (Figure 3) requires an Orion dataset of ligands and SMIRNOFF-format force field files as input. It’s essential that the supplied ligands possess reasonable 3D coordinates, including all atoms, and maintain correct chemistry, including appropriate bond orders and formal charges. Bespoke force field files are in the OFFXML file format. These bespoke force field files only cover specific parameters for the ligand. To complete the force field parameters, integration with a general force field is necessary. As of the present version, the bespoke force field parameters are compatible and can be integrated with OpenFF_2.0.0 and OpenFF_1.3.1.
How to apply bespoke force field
Begin by loading the ligand dataset to the input: ‘Ligand Input Dataset’
Choose the appropriate general force field in the input: ‘Ligand Force Field’
Load the bespoke force field files to the input: ‘Ligand Bespoke ForceField File’
Name the output dataset
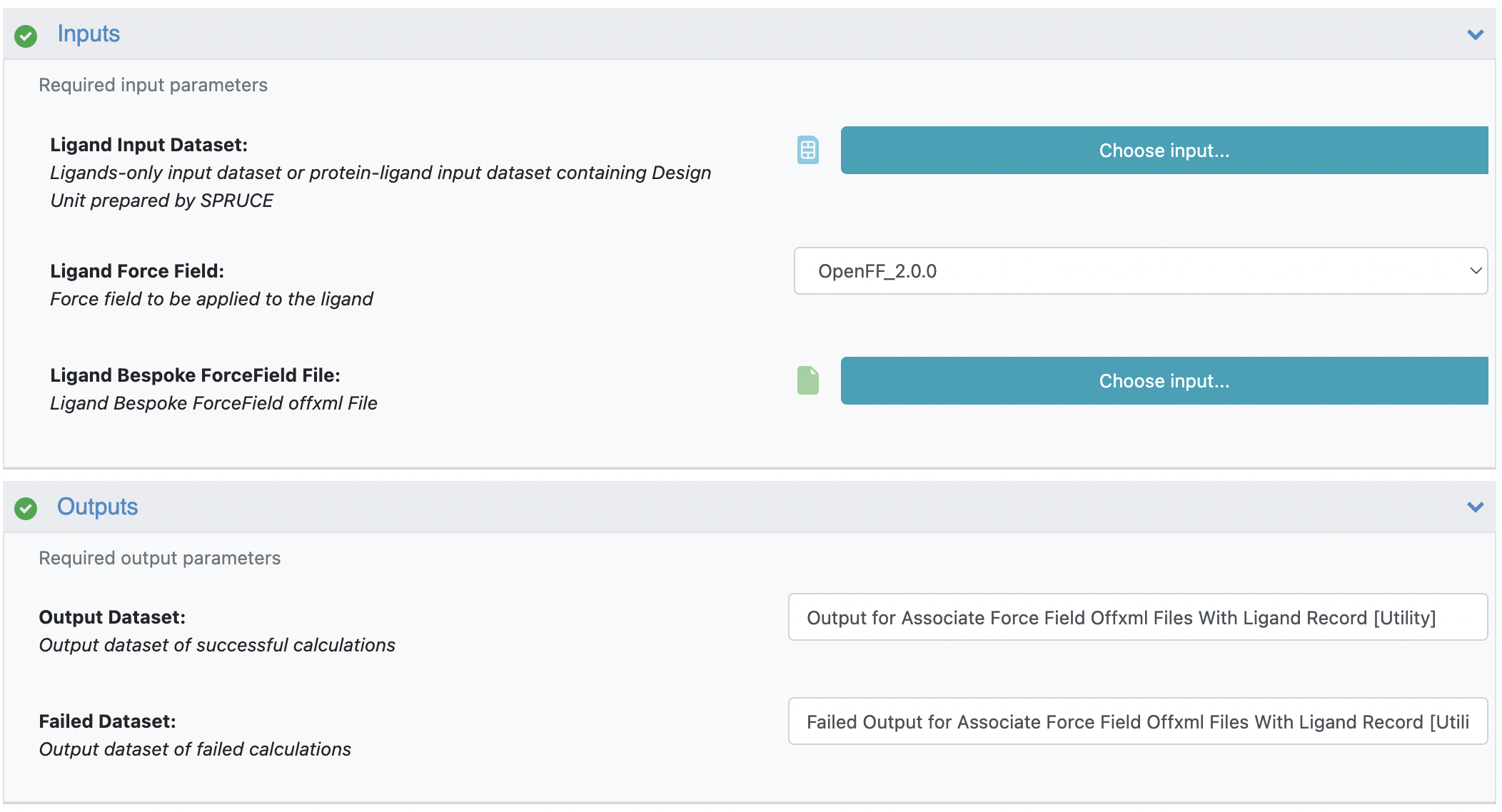
Figure 4: The input and output selections of the floe
The output ligand dataset generated from this floe can serve as input for other
MD floes such as Bound Protein-Ligand MD, Short Trajectory MD with Analysis, or
Ligand Bound and Unbound Equilibration for NES.
By default, when bespoke force field parameters are identified within a dataset,
they are automatically applied to the system.
This automatic application can be disabled if it’s desired.
It can be achieved by switching the setting of Use Bespoke Parameters When Available to Off
in the Apply Force Field cube and Apply Force Field On Unbound States cube if applicable within the unpromoted section.
The unpromoted section can be viewed by turning Show cube parameters to On.
How to apply pre-created custom force field
A custom force field functions as a comprehensive force field that is applicable to the entire ligand dataset, rather than parameterizing a specific ligand. Custom force field files must be in the SMIRNOFF XML format. Below is an example of a custom force field file. This comprehensive force field file includes full sections of parameters. The provided example shows the customization of a specific SMIRKS pattern, [c:4]1cc(c[c:4][c:3]1[c:2]2[c:1]cc(n[c:1]2)N)N, across the entire original force field parameters. When this custom force field file is applied to the flask, the corresponding parameter will be applied if there is a fragment in the ligand that matches the specific SMIRKS in the force field file. This showcases how this file serves as a parent force field applied to the entire system. The highlighted fragment (Figure 5) corresponds to the specific SMIRKS pattern, indicating a targeted force field customization.
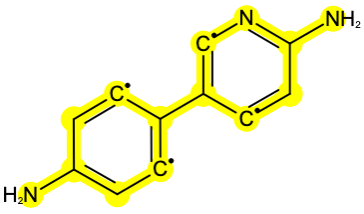
Figure 5: The highlighted fragment matches the SMRIKS pattern from above.
When integrating custom force field, follow these steps (Figure 6):
Adjust the Ligand Force Field setting to
Custom.Upload the custom force field file to the Custom Ligand Force Field File field.
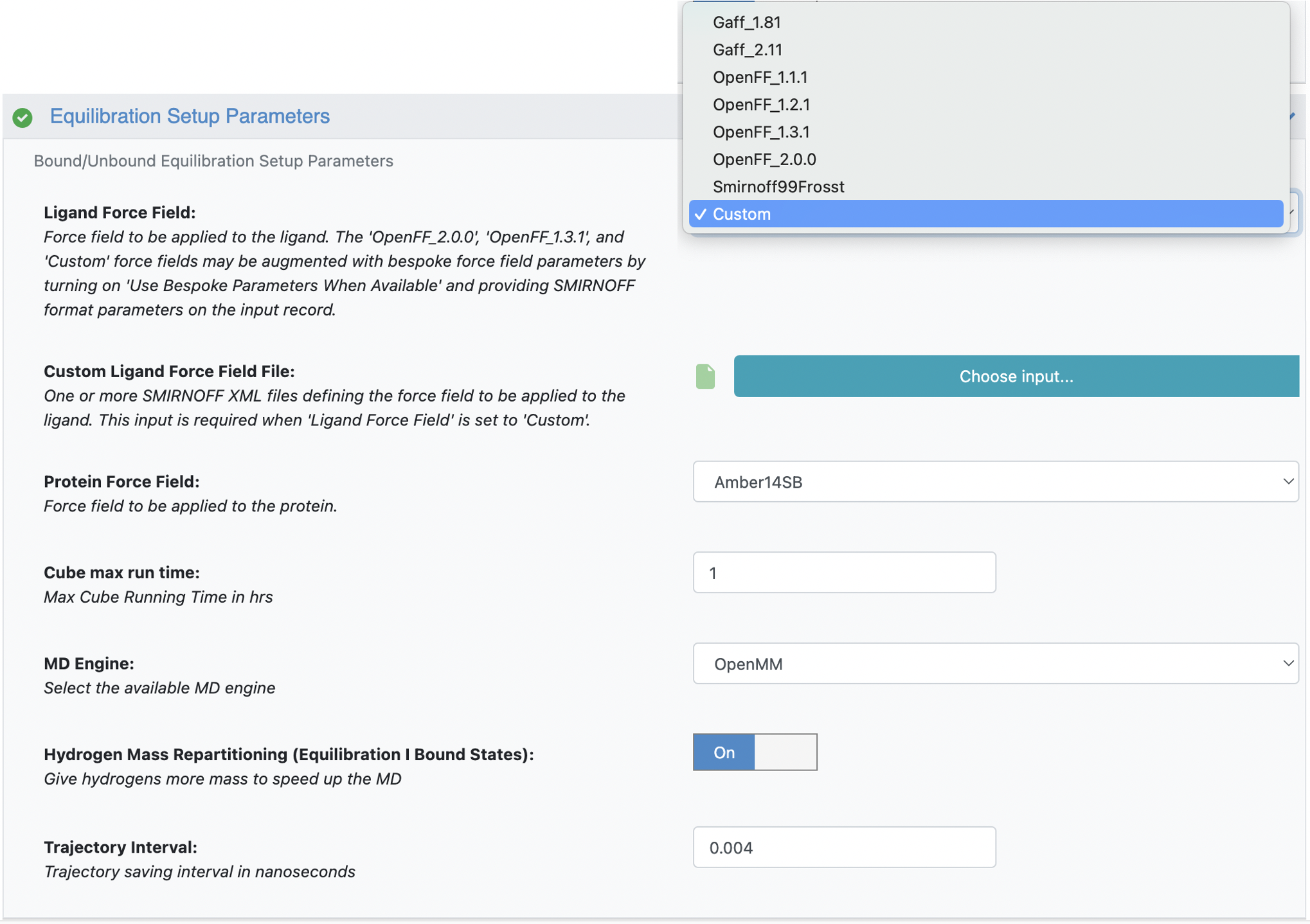
Figure 6: Upload custom force field file
Note
It’s important to note the precedence order of force field parameters: bespoke > custom > general. In the case of identical SMIRKS patterns for the same parameter, the ‘last match wins’ rule determines the application of parameters, meaning that the parameter that appears last will take precedence.
Footnotes