Basic Tutorial: Automated Cryptic Pocket Detection with Probe Occupancy Analysis
Quick floe search term: CPD A1-C2
This tutorial demonstrates use of the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe on KRas. This floe performs mixed-solvent simulations with 150 mM of xenon probes and performs probe occupancy analysis to identify potential cryptic pockets as sites with high xenon occupancy.
In previous versions of the Cryptic Pocket Detection package, this floe was provided as a set of multiple floes (A1-C2) that were run sequentially. You can still run the modular advanced floes sequentially instead of this single floe, starting from the Solvate and Equilibrate Target Protein Floe and going to the Probe Occupancy Analysis Floe. The instructions for doing this are provided in the Advanced Tutorials.
In this section, we will show you how to use the Spruce-prepared KRas protein for the
Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe.
For an example of how to prepare the apo 4OBE protein structure design unit, see the
Preparing Input tutorial.
Alternatively, you can download the prepared design unit for the 4OBE protein structure of
KRas.
KRas 4OBE protein structure DU
Caution
Running the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe instead of the modular advanced floes (A1-C2) incurs an overhead cost. The amount of overhead cost depends on the system size and number of iterations in weighted ensemble MD.
Tip
This floe typically takes more than a day to finish and costs ~$500.
Search and Run the Floe in Orion
Locate the Floe in Orion
Navigate to the Floe page using the blue navigation bar.
On the Floe page, click on the Floes Tab, where you will find a list of the available floes and packages.
Under the Category Floe Filters on the left, click on the caret next to the Packages filter to expand the list of packages and click on the OpenEye Cryptic Pocket Detection Floes package. This will ensure that the floes listed in the middle of the page are from this package.
From this list, click on the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe, and then click the blue “Launch Floe” button to launch the Job Form, as shown in Figure 1.
Provide Input Files and Parameters to Run the Floe
- Output path:
On the Job Form, specify where your output data should be directed.
- Input Data:
Choose the Target Protein to be the Spruce-prepared dataset for KRas.
- Output Data:
You can customize the output dataset and collection names here.
- Protein Solvation And Equilibration Advanced Settings:
Advanced parameters for solvation and equilibration of the target protein can be set here. Click on the caret to expand this section and display the parameters. Additional details can be found in the floe description of the Solvate and Equilibrate Target Protein Floe. These parameters do not allow changing the mixed-solvent condition, which is set to 150 mM xenon. For more control over the solvent composition, we recommend running the modular advanced floes.
- Normal Modes Calculation Advanced Settings:
Advanced parameters for normal mode calculation can be set under this option. Click on the caret to display a list of parameters that can be adjusted if required. Additional details on these parameters can be found in the Calculate Normal Modes Floe documentation.
- Weighted Ensemble MD Advanced Settings:
These advanced parameters allow you to change the total number of iterations and other weighted ensemble parameters. Additional details on these parameters can be found in the Run a Normal Mode-Guided Weighted Ensemble MD Simulation Floe documentation.
- Weighted Ensemble MD Analysis Advanced Settings:
Advanced settings for performing weighted ensemble trajectory analysis are provided under this section. See the Perform Weighted Ensemble MD Analysis Floe for additional details on these parameters.
- Cryptic Pocket Analysis Advanced Settings:
Functionally important residues, such as active site residues or known disease mutations, can be provided as input for the Important Residues parameter. These residues will be displayed along with cryptic pocket residues in the cryptic pocket analysis Floe Report. See the Probe Occupancy Analysis Floe documentation for additional details.
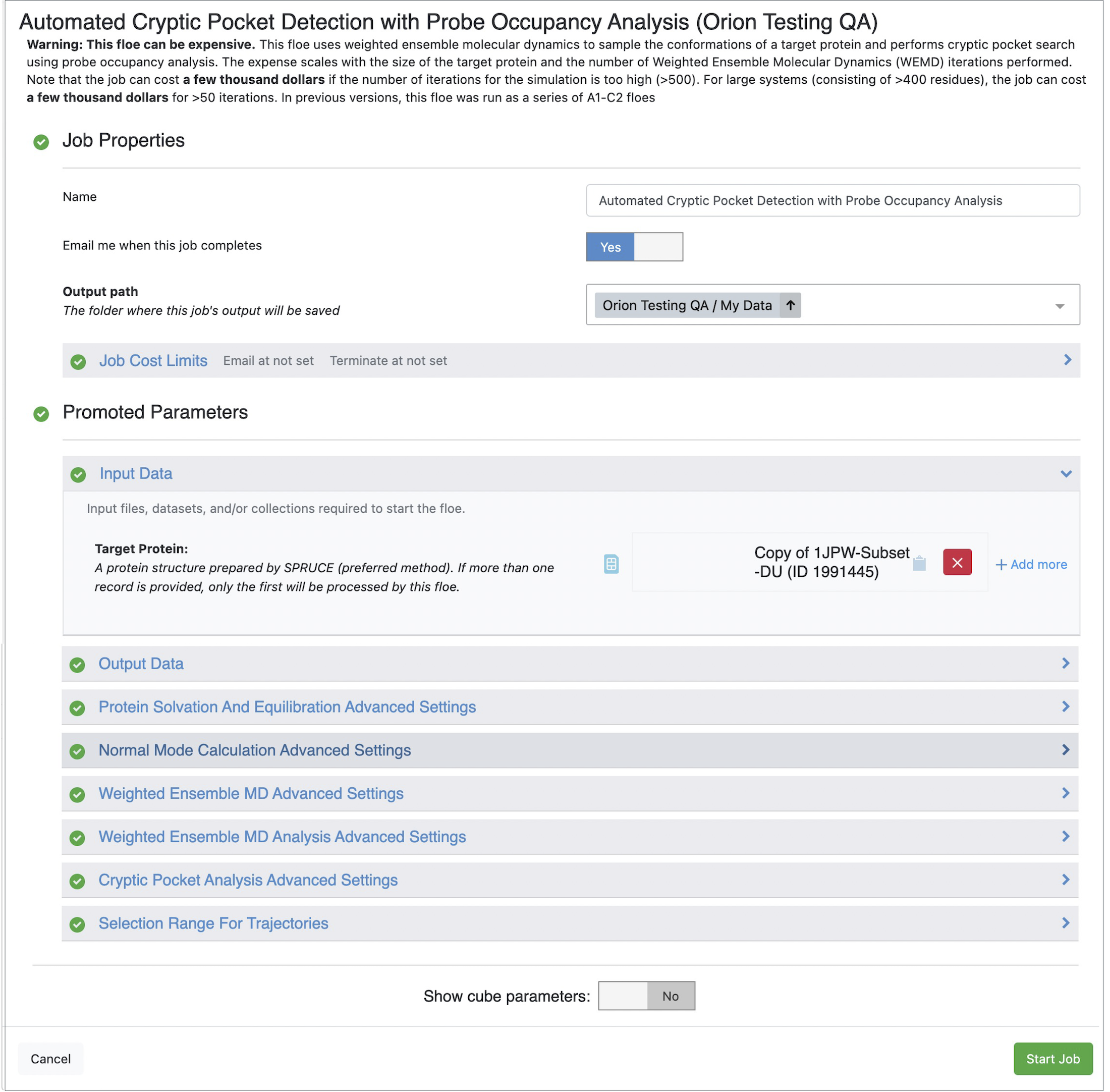
Figure 1. Job Form for the Automated Cryptic Pocket Detection with Probe Occupancy Analysis Floe.
After providing the input design unit and optionally adjusting input parameters, click on the green “Start Job” button.
Visualize Cryptic Pocket Analysis Report and Pocket Receptors
Cryptic Pockets Floe Report (Probe Occupancy Analysis)
Access the Floe Report.
When the job is complete, the output Floe Report, Cryptic Pockets Floe Report (Probe Occupancy Analysis), should be inspected for visualization of cryptic pockets. To reach the Floe Report, navigate to the Jobs Tab on the Floe page and then click on the job that you want to inspect. Under Reports, click on the Floe Report Cryptic Pockets Floe Report (Probe Occupancy Analysis). This will redirect you to a report containing an interactive network plot of pockets (Figure 2) detected as high-occupancy probe binding sites.
Visualize the interactive network plot.
Each node in the interactive network plot represents a pocket, and the edge connecting two pockets corresponds to the inverse of the center-of-mass distance between two pockets. Node size corresponds to the probe occupancy free energy. The range of node colors corresponds to the number of pocket residues. By clicking on a node, a visualization of a representative protein configuration appears with the pocket-forming residues highlighted by a blue surface. If the Important Residues parameter is provided by the user, those residues will be highlighted by a pink surface along with pocket-forming residues. You can visualize the residue side chains by clicking on the “Show Residues” button given at the bottom left corner of the page. Alternatively, clicking on an individual residue atom will show the label for that atom. Hovering over an edge in the network plot will display edge metadata.
Download ranked pockets data.
You can also download the metadata for the ranked pockets by clicking on the “RankedPockets.json” link in the Download Figure Data field. This file lists ranked pockets, their residue composition, and probe occupancy free energy.
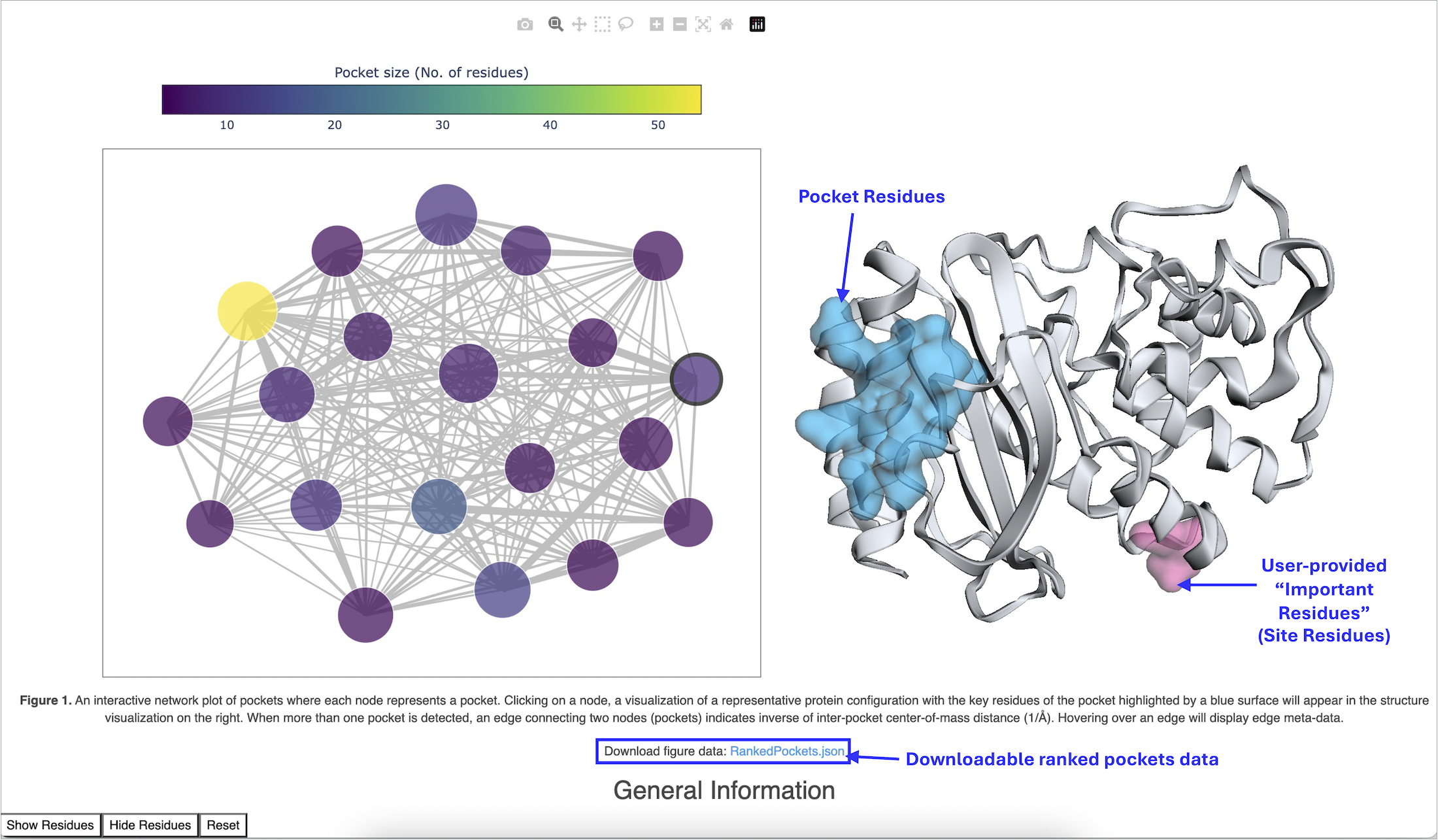
Figure 2. An interactive network plot of cryptic pockets.
Pocket Receptors (Probe Occupancy Analysis) Dataset
Access the pocket receptors dataset.
After the job is complete, you can get to the details of your job (in this case, the dataset Pocket Receptors (Probe Occupancy Analysis)) by clicking on the job. Navigate to the Jobs Tab on the Floe page. Click on the job that you want to inspect. Click on the “View in Project Data” button next to ‘Results.’ This will redirect you to the Data page and show only the outputs associated with the job. Next to the dataset (default name: Pocket Receptors (Probe Occupancy Analysis)), click on the circle with a + sign to activate the dataset. It will change to a green circle with a checkmark and will allow you to view the dataset in the 3D & Analyze page.
Visualize the pocket receptors dataset in the 3D & Analyze page.
Using the navigation bar, go to the 3D & Analyze page. Make sure that your Active Dataset is set to your Pocket Receptors (Probe Occupancy Analysis) dataset. On the scatter plot on the 3D & Analyze page, you can choose Normalized Ligandability Score for the y-axis and Pocket Rank for the x-axis. Also click on the ‘Layouts’ drop-down in the Active Data Bar and select the 3D Analyze view to visualize a design unit with a pocket receptor. This shows a depiction of the protein structures of the representative conformations with a receptor corresponding to a selected pocket and normalized ligandability score of the pocket.
The Pocket Rank column in the Spreadsheet shows the pocket rank determined by normalized ligandability score of a pocket. The pocket rank 1 has the highest predicted ligandability.
The Spreadsheet shows the normalized ligandability score, ligandability score per receptor volume, total pocket ligandability score, ligandability score significance, and receptor volume for a pocket in a representative conformation selected from the cluster center conformations generated during cryptic pocket analysis. The pocket representative conformations is selected for each pocket (high occupancy probe binding site) based on the normalized ligandability score values for receptor volumes within the rage of 500 to 2100 Å3. Users can change the receptor volume range used for the selection of the most ligandable conformation for a pocket.
The Reference Receptor Volume column in the Spreadsheet shows the receptor volume for a pocket in the equilibrated structure used to start the weighted ensemble MD simulation. Comparison of this value with the Receptor Volume value provides an indication of the pocket opening and closing during the simulation.
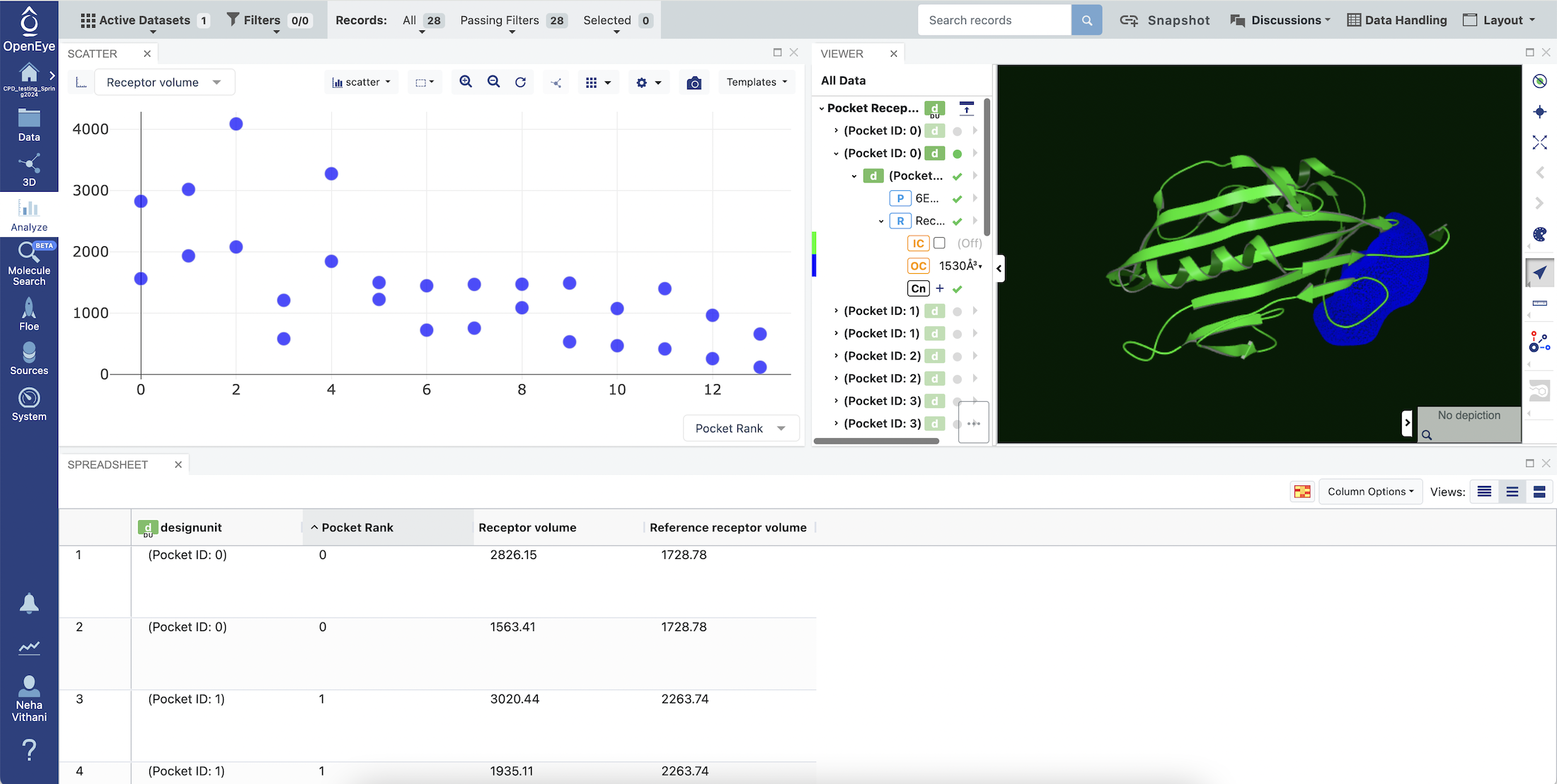
Figure 3. Pocket Receptors (Probe Occupancy Analysis).
Sort and Select Pocket Receptors.
Clicking on the Pocket Rank column in the Spreadsheet Panel sorts the representative conformations by the pocket rank in these conformations, in either ascending or descending order.
After sorting the structures by rank in the Spreadsheet, click on a row with the Pocket Rank of choice. This will display the protein structure in the Viewer panel corresponding to the selected row.
Under All Data, click on the small caret next to the corresponding design unit to display all components present in this design unit.
Click on Receptor, Inner Contour (IC), and Outer Contour (OC) to visualize the receptor. The receptor will appear in blue-colored mesh. After visualizing different design units and their receptors, you can select an appropriate design unit for Gigadock or SiteHopper analysis.
Failure Report
Your job might finish with the job status “Success” and generate a Failure Report, if no cryptic pockets are detected by this floe. Open the Failure Report to see the instructions. The analysis can fail to detect cryptic pockets for multiple reasons.
The cryptic pocket detection method you chose failed to detect a pocket. It is possible that one or all of our cryptic pocket detection methods fail to detect the pockets. All three methods use different approaches and define “cryptic pockets” in a different manner. For example, the Probe Occupancy Analysis Floe will fail to detect pockets if no sites with high probe occupancy were identified.
No significant conformational changes associated with cryptic pocket formation were observed during the simulation. This could happen because of insufficient sampling or when the normal modes used as progress coordinates could not efficiently sample pocket formation. You may consider extending your weighted ensemble MD simulation using the Continue a Normal Mode-Guided Weighted Ensemble MD Simulation Floe and rerun the cryptic pocket analysis with the extended protein sampling. Alternatively, you can perform another weighted ensemble MD simulation using a different set of normal modes as progress coordinates with high variance in the region of interest in the target protein.
It is also possible that your target protein is highly inflexible; therefore, it doesn’t show conformational changes that can potentially reveal a cryptic pocket.