Induced-Fit Posing (Confined) How-To Guide
What this Floe does
The Floe Induced-Fit Posing (Confined) performs Induced-Fit Posing (IFP). Given a reference receptor and a ligand to dock, it carries out the following three steps:
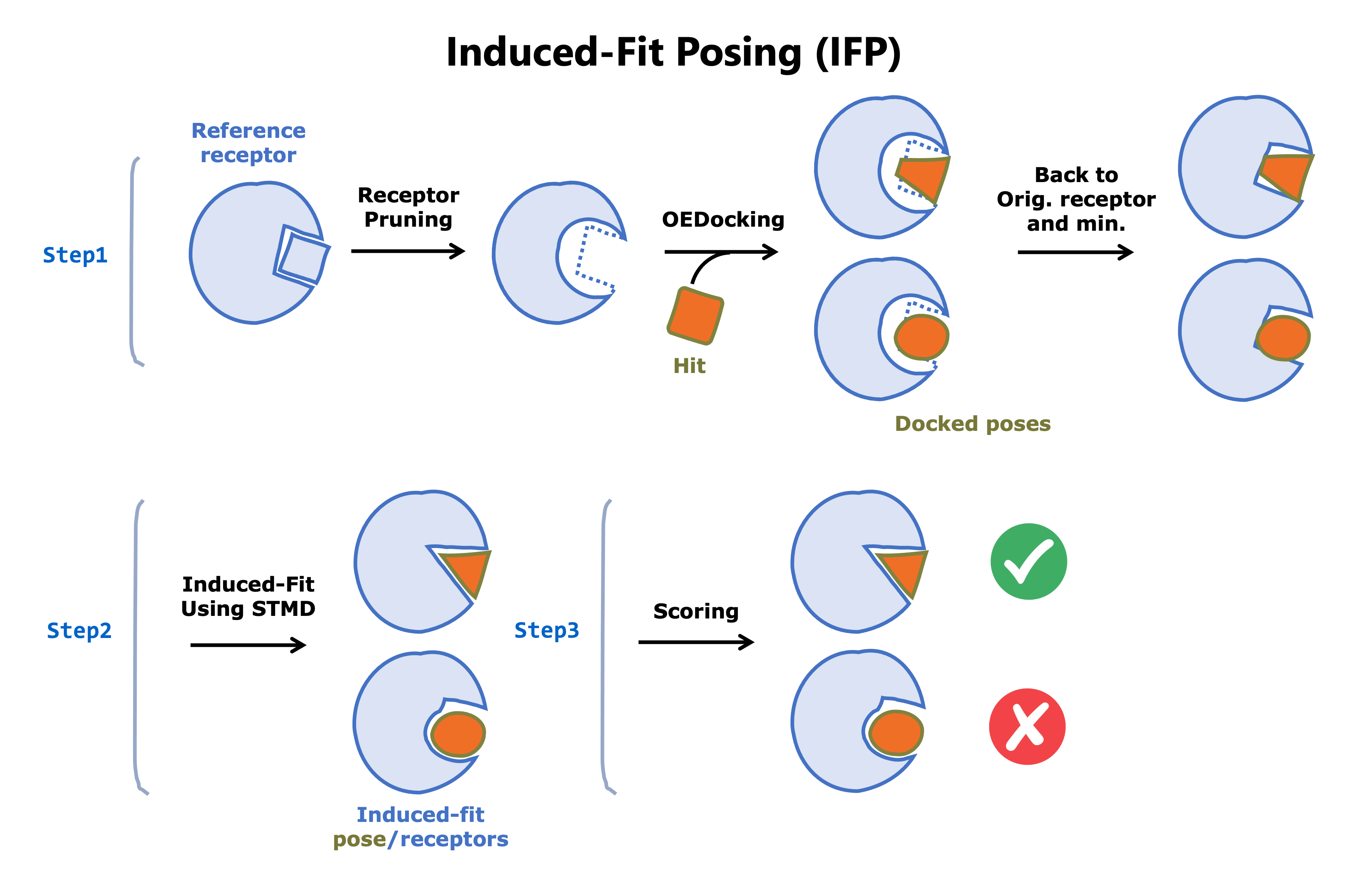
Docking and initial processing of docked pose/receptor conformations
OMEGA conformer ensemble generation of the ligand to be posed
Pruning binding pocket residues in the reference receptor to alanine
Docking of the ligand to both pruned and un-pruned receptors followed by initial scoring to select the top 12 pose/receptor conformations
Molecular dynamics (MD) simulations for induced-fit between the receptor and the posed ligand followed by trajectory analysis and extraction of the induced-fit pose/receptor conformation from the trajectory.
Final scoring (IFP Score) of the induced-fit pose/receptor conformations. Higher IFP scores represent greater confidence that the predicted induced-fit conformation is accurate.
We refer to the induced-fit process as confined due to its limited scope. The output datasets can be used as input datasets for other MD Floes for further sampling, etc.
Scope and limitations
The input reference protein dataset must contain a bound ligand (An apo state of the reference is not supported in this Floe). The bound ligand of the reference protein will not appear in the final results but will be used to guide the initial docking of the compound provided in the docking ligand input dataset.
The Floe takes a single reference receptor and a single docking ligand at a time.
Limitations of the current implementation: (1) the induced-fit procedure may not sufficiently reorganize protein secondary structure, tertiary structure, or loop regions where backbone rearrangement is necessary; (2) reference proteins with small outer contours relative to the docking ligand may suffer during docking. To address this issue, see Tips for the users
Input requirements
The ligand to dock needs to have reasonable 3D coordinates, all atoms, and correct chemistry (in particular, bond orders and formal charges).
The reference receptor needs to have a unique OEReceptor and needs to be prepared to MD standards: protein chains must be capped, all atoms in protein residues (including hydrogens) must be present, and missing protein loops resolved or capped. Typically, SPRUCE is used for protein preparation.
How to run this Floe
After selecting the Induced-Fit Posing (Confined) Floe in the Orion UI, you will be presented with a job form with parameters to select. Aside from the required input (Ligand Input Dataset and Reference Receptor Input Dataset), all other parameters have reasonable defaults.
The optional inputs are:
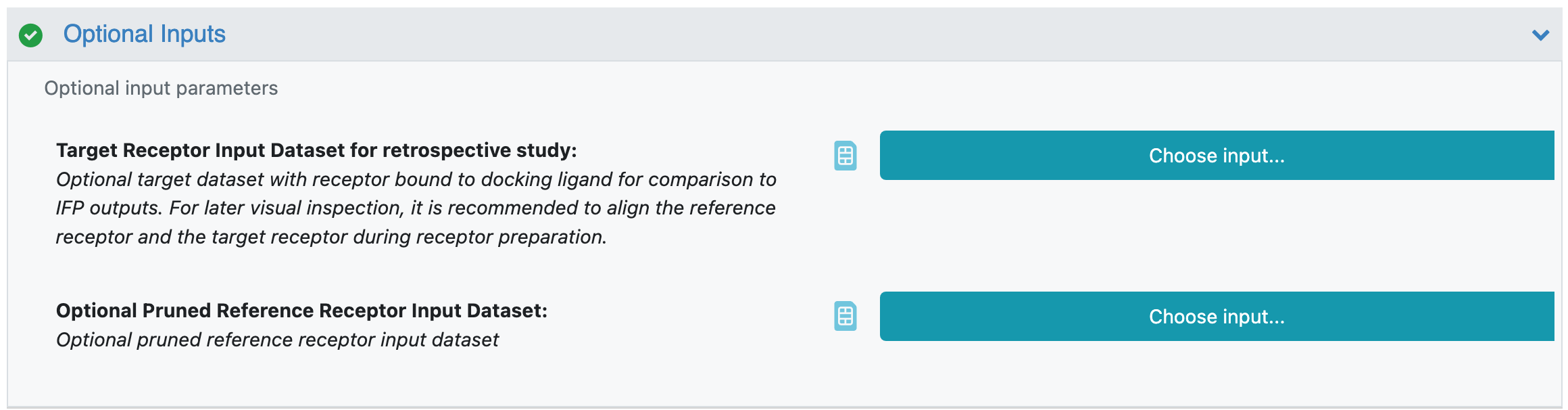
Target Receptor Input Dataset for retrospective study (no default): Optional target receptor with the known bound ligand for comparison to IFP outputs. If the target receptor which has the same binding site residues as the reference receptor is provided, pose RMSD to target pose, active site side chain RMSD to the target receptor, and active site side chain torsion fingerprint deviation (TFD) to the target receptor will be computed. For later visual inspection, it is recommended to align the reference receptor and the target receptor during receptor preparation.
Optional Pruned Reference Receptor Input Dataset (no default): Optional pruned reference receptor to use in docking. One can generate a pruned receptor input manually or by using the Induced-Fit Posing Prep: Pruning reference receptor for docking Floe. To apply constraints on the pruned receptor input, one can (1) define and activate constraints to force during docking using OpenEye’s MakeReceptor GUI desktop application, OpenEye’s ReceptorToolbox utility program, or via Orion’s 3D modeling page; (2) then input the customized pruned receptor with activated constraints to Optional Pruned Reference Receptor Input Dataset. If the optional pruned reference receptor is given, the Floe does not proceed with receptor pruning and uses the input pruned receptor instead.
The top-level parameters for Docking:
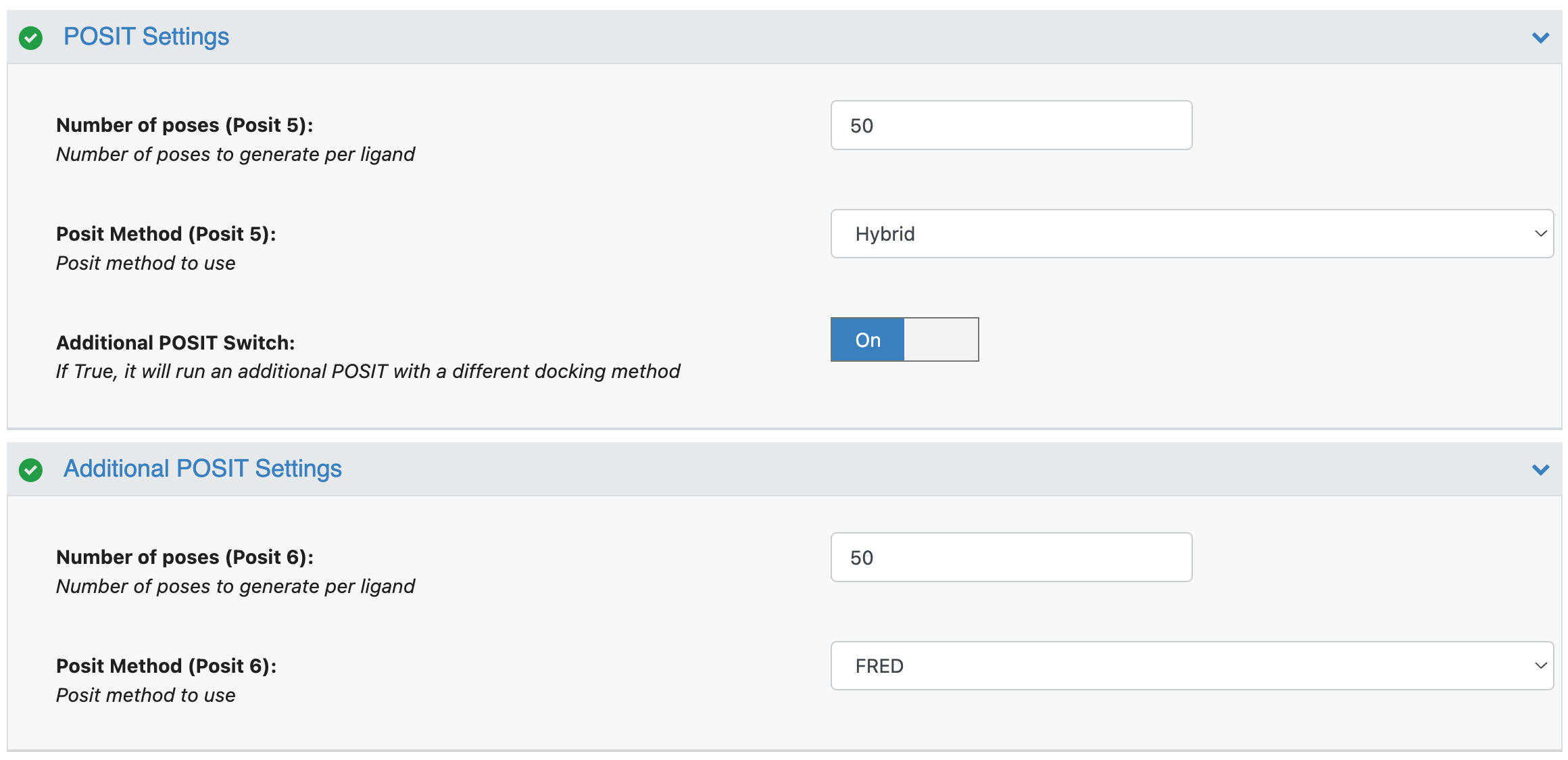
POSIT Settings/Number of poses (default 50): Number of poses to generate per receptor
POSIT Settings/Posit Method (default Hybrid): Docking method to use
POSIT Settings/Additional POSIT Switch (default On): If On, it will run an additional Posit with a docking method selected in Additional POSIT Settings/Posit Method.
Additional POSIT Settings/Number of poses (default 50): Number of poses to generate from an additional Posit per receptor
Additional POSIT Settings/Posit Method (default FRED): Docking method to use for additional Posit
We make the other top-level parameters available for expert users by turning on Show Cube Parameters at the bottom of the input form and then drilling down into the parameters of the desired Cube in the list below.
Tips for the users
If the cognate ligand of the reference receptor is much smaller than the ligand to be posed, the outer contour of the default OEReceptor has a chance to be too small to allow enough space to correctly place poses during docking. For this case, we recommend modifying the OEReceptor outer contour using OpenEye’s MakeReceptor GUI desktop application or via Orion’s 3D modeling page.
How to check results
The results from the IFP Floe are accessed via two main avenues: (1) through the job output in the Jobs tab on Orion’s Floe pages; (2) through Orion’s 3D and Analyze pages.