Rapid FE-NES
Free Energy Nonequilibrium Switching (FE-NES) provides a faster, more scalable approach for predicting small-molecule binding affinities using RBFE computations. By replacing slow equilibrium simulations with rapid, parallel transitions, FE-NES delivers accurate free energy predictions at speed as detailed in our recent article. This enables project teams to explore broader design spaces and generate promising hypotheses earlier in lead optimization. Because each alchemical switch is independent, FE-NES is capable of fault-tolerant elastic scaling on cloud resources, making it an ideal fit for the architecture of Orion. Additional details about FE-NES are available in the literature ([Hummer-2001]; [Hummer-2002]; [Gapsys-2020]) and in the general FE-NES tutorial.
Rapid FE-NES (Rapid Free Energy Nonequilibrium Switching) builds on these strengths by amplifying both speed and scale without substantially compromising accuracy. These benefits are achieved not by making the alchemical switches faster but by running fewer of them. As a result, Rapid FE-NES delivers even higher throughput and faster turnaround so that you can schedule synthesis sooner, run more lead optimization cycles, and maintain project momentum. Specifically, Rapid FE-NES can double prediction throughput or triple analog coverage, with a typical increase in prediction error of only 0.2 kcal/mol.
How to Run Rapid FE-NES on Orion
This tutorial demonstrates how to run Rapid FE-NES with the Equilibration and Nonequilibrium Switching Floe.
Quick Reference
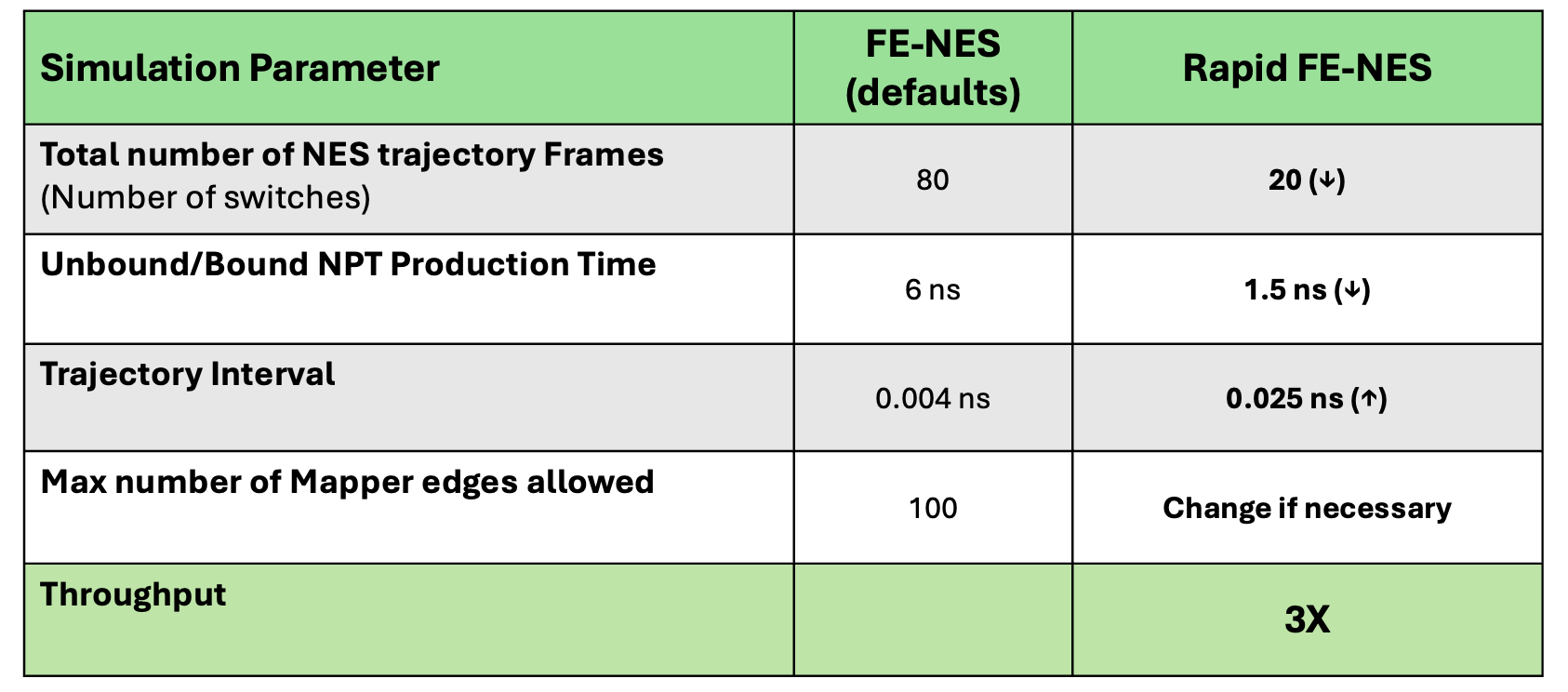
Running Rapid FE-NES with the Equilibration and Nonequilibrium Switching Floe requires configuring three simulation parameters, which are described in this table. These parameters can increase throughput by 3 times compared to FE-NES run with default settings.
Floe Inputs
Three inputs are required, along with one optional input:
- One receptor
- A dataset containing the protein target.Only one protein target (i.e., 1 record) per dataset is allowed. The protein target may consist of multiple chains.The receptor should be prepared with the SPRUCE - Protein Preparation Floe, as described in the Spruce tutorial.
- Multiple ligands
- A single dataset containing at least two ligand records.Ligands must be preposed in the receptor’s binding site. Ligands must be small molecules.Because this floe computes relative binding free energies using a chimeric common core, all ligands must belong to the same congeneric series and share a conserved binding mode.
- One edge map
- A dataset that defines the chemical transformations to be evaluated by FE-NES.This is a graph in which each ligand (node) is connected by an alchemical transformation (edge) to at least one other ligand.Edge map construction is described in more detail in the Edge Mapper tutorial.
- (Optional) Experimental binding affinities
A file listing known experimental affinity values for the ligands (e.g., an uploaded
.txtfile) with the following syntax:[ligand_name] [Affinity] [Error{optional}] [kcal/mol|kJ/mol]
You can complete this tutorial using your own input files. For convenience, we also provide representative inputs below. When viewing links to other tutorials, you may right-click and open the link in a new tab to avoid leaving this page.
Input |
File |
Relevant Tutorials |
|---|---|---|
Receptor
Dataset
|
TYK2 (PDB ID: 4GIH)
|
|
Ligand
Dataset
|
Poses from [Wang-2015]
|
|
Edge Map
Dataset
|
||
Experimental
Affinities
(Optional)
|
Determined from data in [Wang-2015]
|
Locate and Launch the Floe
On the Orion interface, click the “Floe” button on the blue navigation bar to reach the Floe page.
Click on the Workfloes tab.
Under the Floe Filters in the left-hand pane, click on the ‘Packages’ drop-down to expand the list of packages, then select the OpenEye MD Affinity Floes package.
A list of the MD Affinity floes will now be visible in the center panel. Select the Equilibration and Nonequilibrium Switching Floe.
Alternatively, in the center panel, begin typing Equilibration and Nonequilibrium Switching in the search bar until you see that floe appear. Then click the “Launch Floe” button to bring up the Job Form.
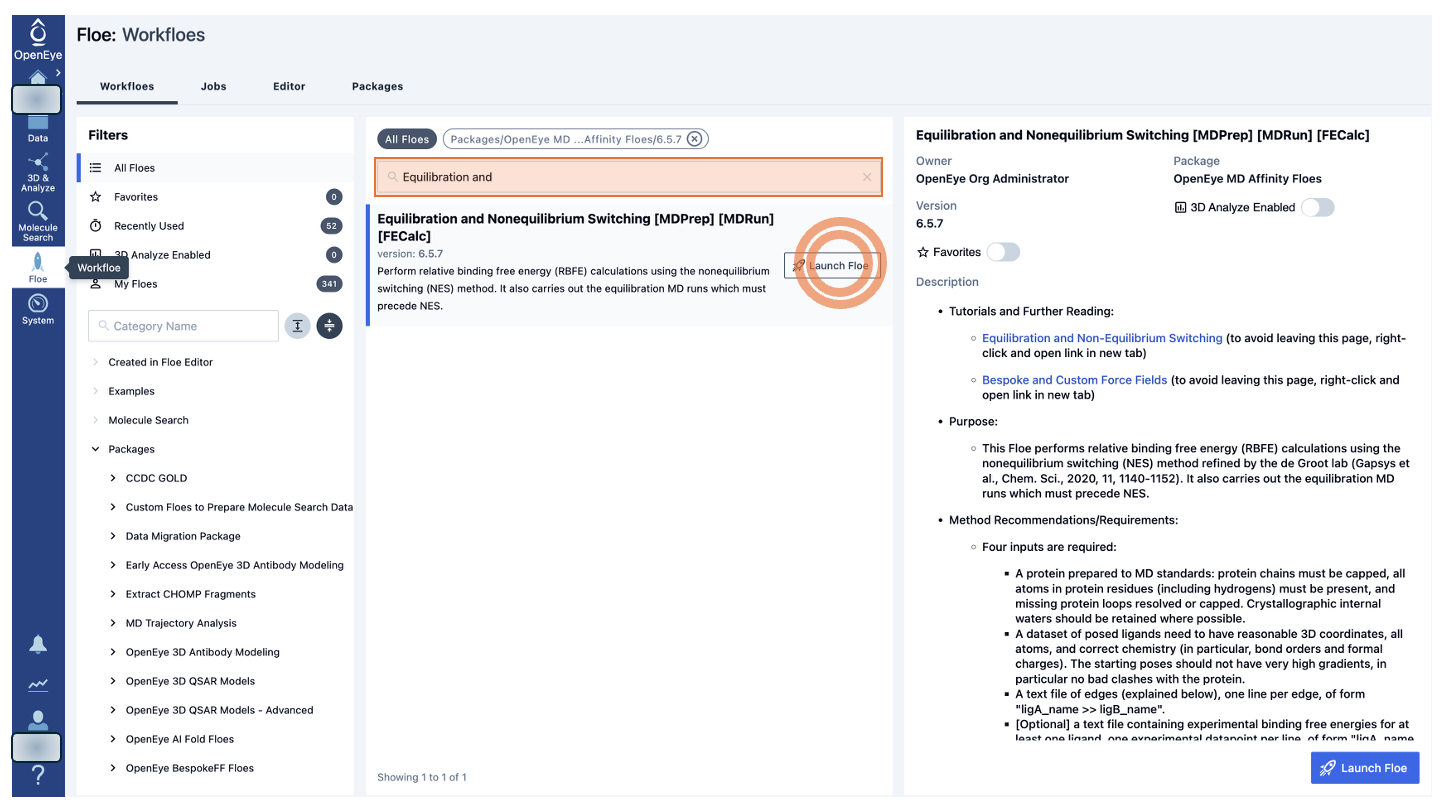
Figure 1. Finding the Equilibration and Nonequilibrium Switching Floe on Orion.
Define Floe Inputs and Submit the Job
Provide the following inputs in the Job Form.
Use the receptor dataset as input for Protein Input Dataset.
Use the ligands dataset as input for Ligand Input Dataset (Ligand Reader).
Use the edge map dataset as input for Mapper Dataset (NES Mapper Reader).
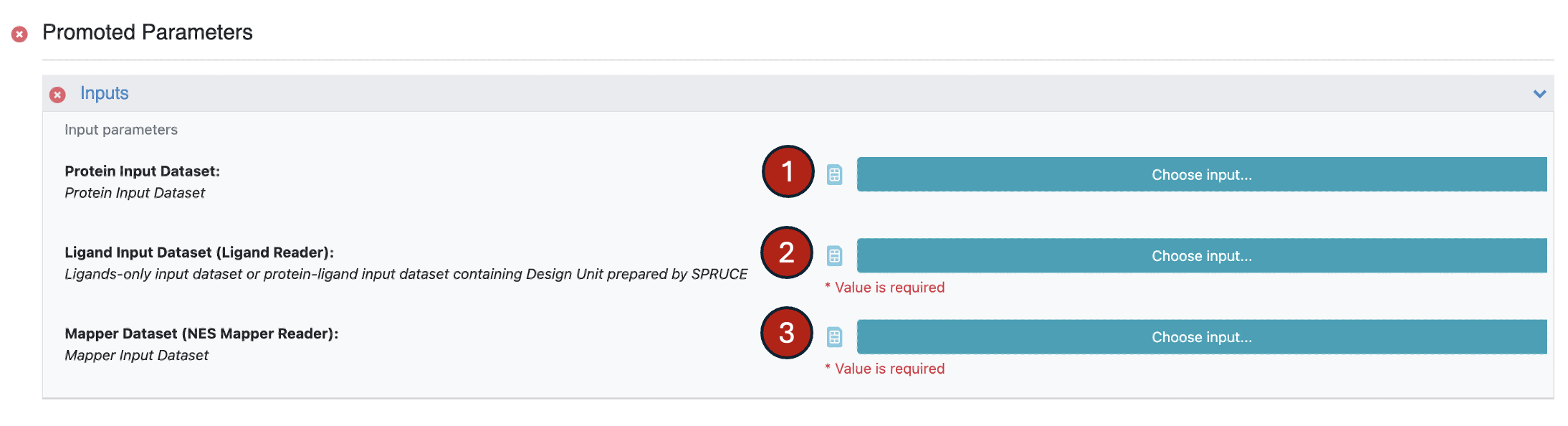
Figure 2. Inputs for the Equilibration and Nonequilibrium Switching Floe.
Optional, but recommended for all retrospective studies: For the Experimental Affinity parameter, provide a file with known experimental binding affinity values for the ligands.
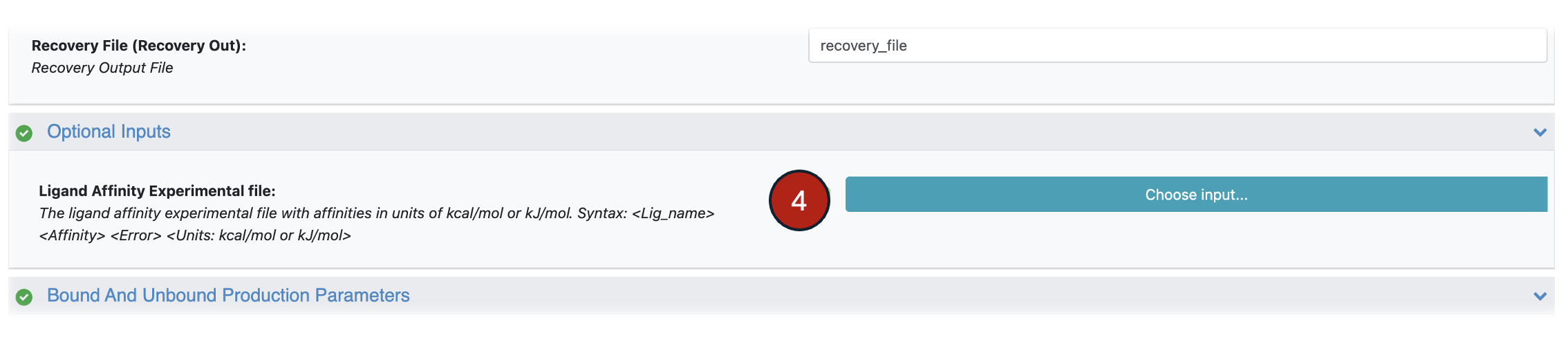
Figure 3. Input for the Ligand Affinity Experimental file.
Rapid FE-NES parameters
To run Rapid FE-NES, you must first configure three additional simulation parameters.
If you skip these parameter modifications, the floe will launch using default FE-NES settings.
In the Bound and Unbound Production Parameters group, under the Bound/Unbound States NPT Production Runtime parameters, change the simulation production times of the bound and unbound states to 1.5. This four times reduction from the default settings helps the end-state simulations to finish faster.
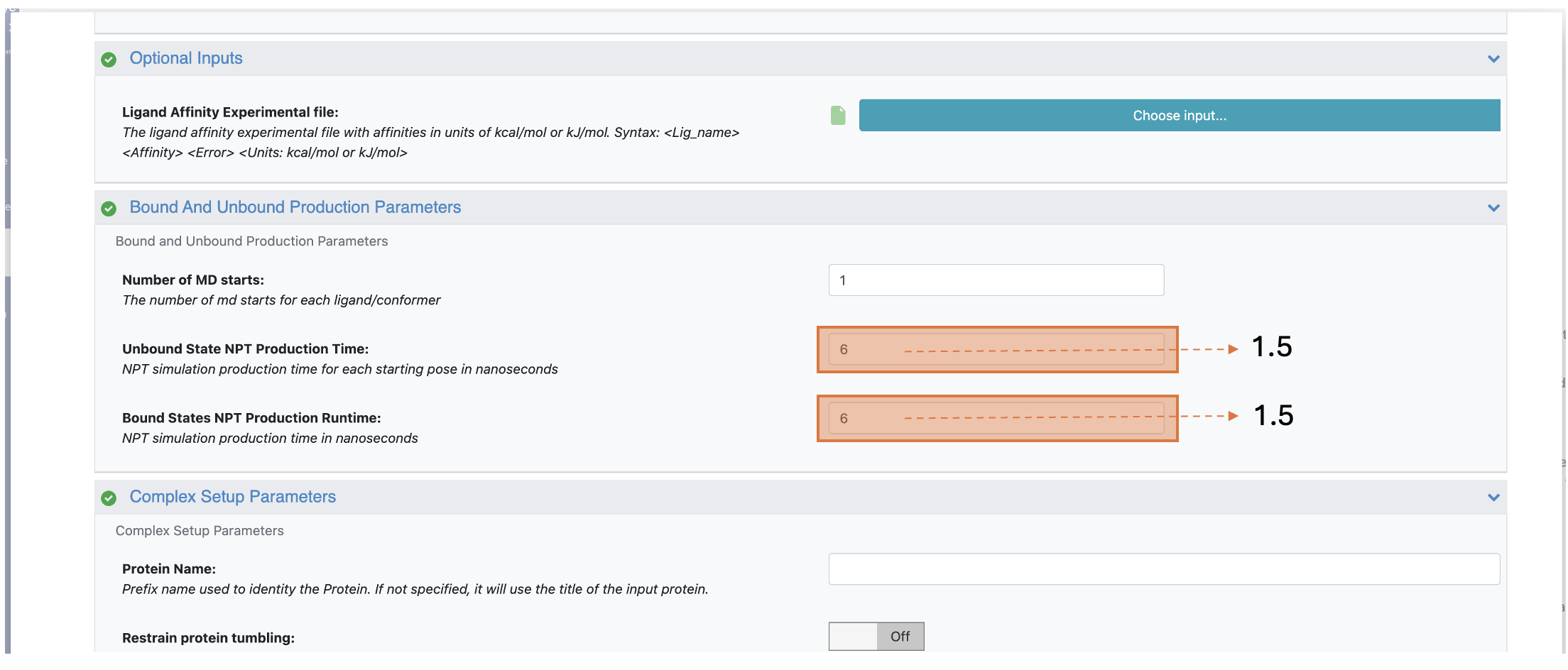
Figure 4. Changing the Production Simulation Times parameter to reduce simulation time.
In the Equilibration Setup Parameters group, change the Trajectory Interval to 0.025. This six times increase from the default settings reduces storage costs and increases overall floe throughput.
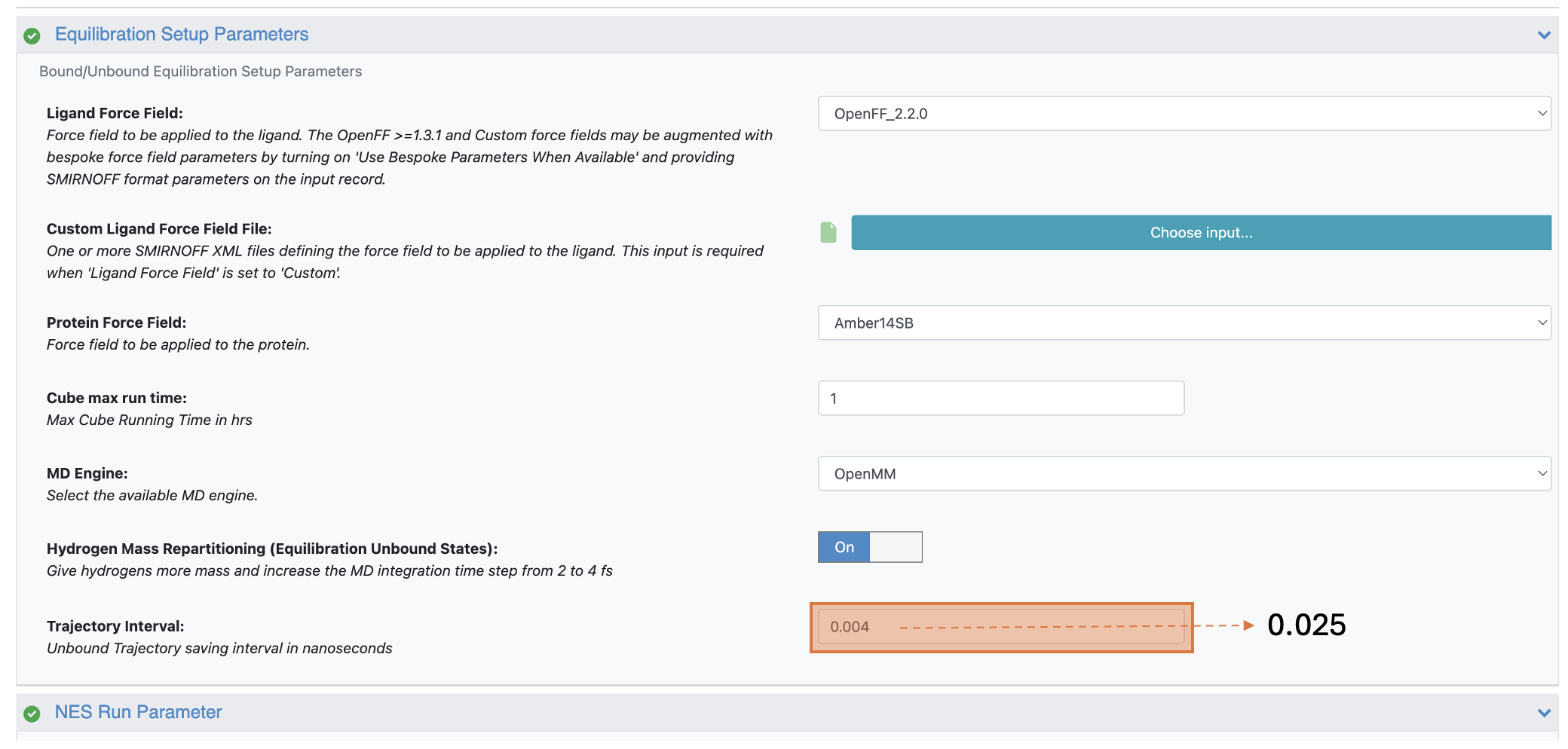
Figure 5. Changing the Trajectory Interval parameter to reduce storage costs and improve floe throughput.
In the NES Run Parameter group, change the Total Number of NES Trajectory Frames to 20. This four times reduction from the default settings helps the alchemical switches finish faster.
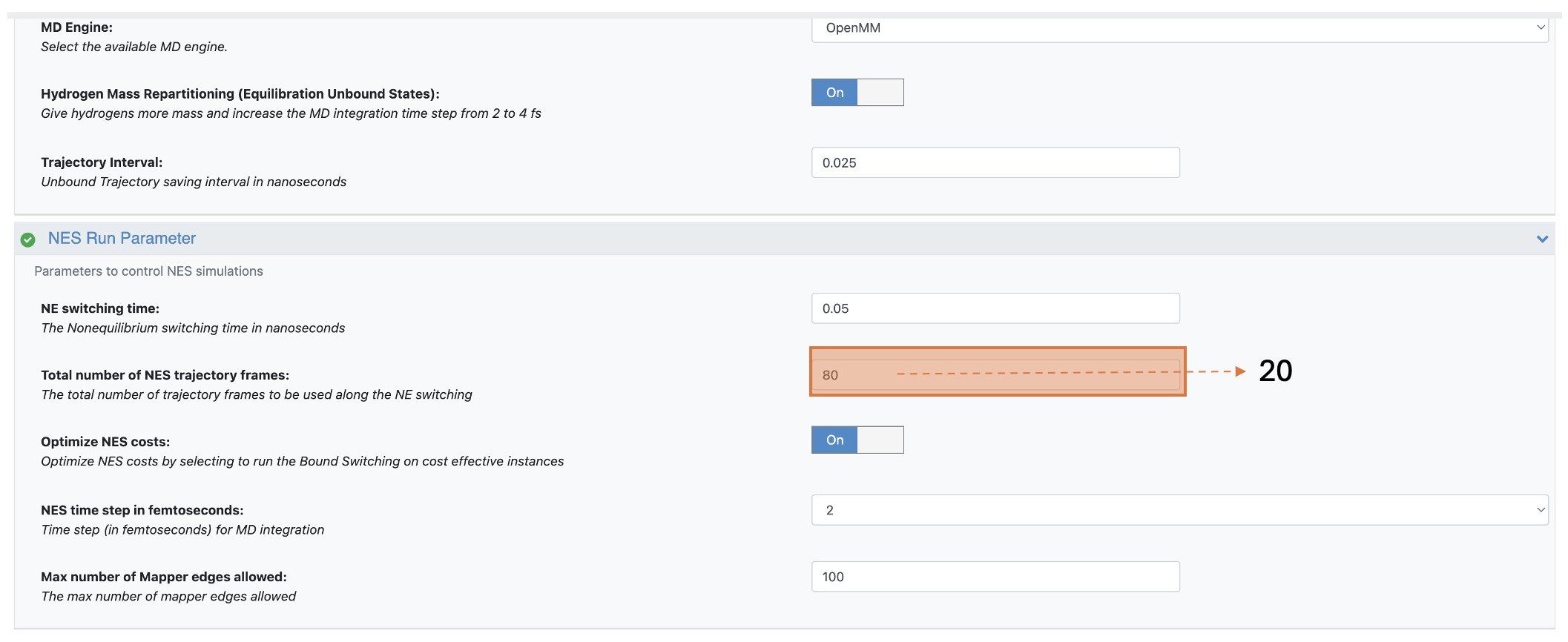
Figure 6. Changing the Total Number of NES Trajectory Frames parameter to speed up the alchemical switches.
Warning
If you are using your own input files, you may need to adjust the Max Number of Mapper Edges Allowed in the NES Run Parameter group. This value defaults to 100 to prevent unintended submission of high-cost jobs. When submitting larger computations, set this value higher than the number of edges you intend to run.
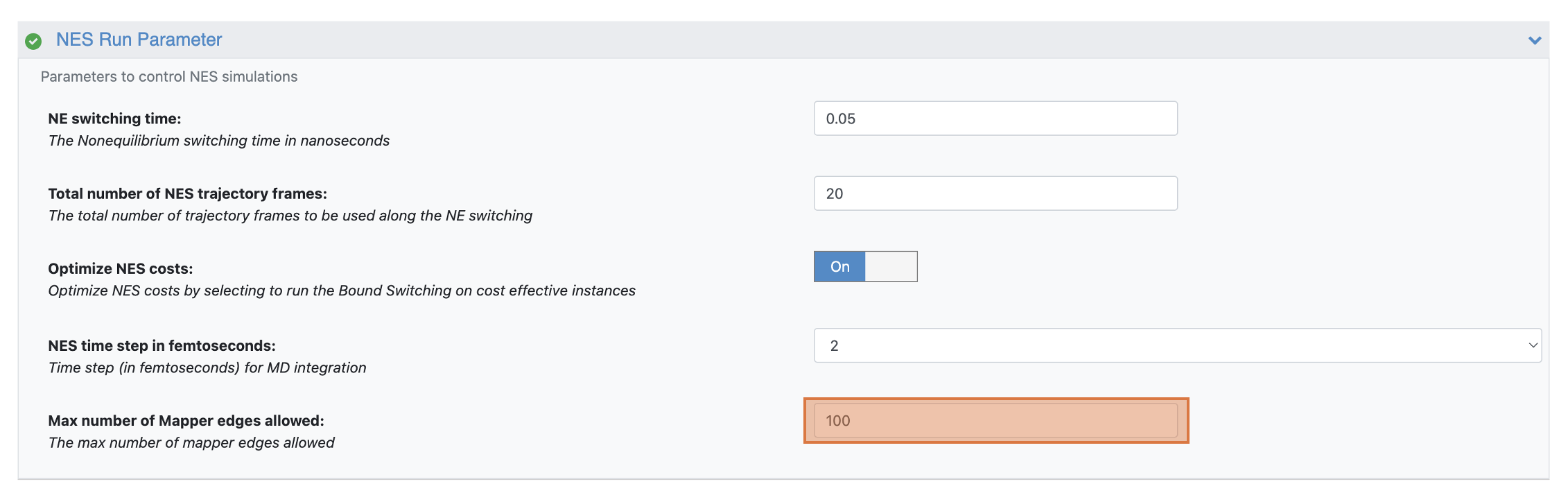
Figure 7. Setting the Max Number of Mapper Edges Allowed parameter to 100 prevents the unintended submission of high-cost jobs.
Click “Start Job” to begin the floe.
Success
Your job has been successfully submitted! Estimated completion time: 2–4 hours.
This estimate is based on the inputs provided in the tutorial, that is, 16 ligands connected by 22 edges. Completion times may vary depending on the size of the input system.
Monitoring Progress
You can track job progress in real time by monitoring the Floe map. On the Floe page, go to the Jobs Tab, then click on your job. For more information, see the the Floe Visualization section in the Floe page of the Orion User Guide. Complete features of the Jobs tab can also be found in the Orion User Guide.
Analyzing Outputs
The Equilibration and Nonequilibrium Switching Floe produces several useful outputs. For more information on their interpretation, see the Understanding the Results from NES section of the Nonequilibrium Switching tutorial.
Standardizing a Floe for Rapid FE-NES
You can save a copy of the floe with the reconfigured simulation parameters for Rapid FE-NES as defaults using the Floe Editor and share it with other users. This will help standardize the workflow across teams. For questions or assistance, please contact OpenEye Customer Support at support@eyesopen.com.