Tutorial: Prepare Supporting Experimental pKa Data File
Gaussian Process Regression (GPR) is a non-parametric regression technique that carries all training data along with optimized parameters to predict pKa values. The floes in this package use a GPR model with descriptors which are optimized for pKa prediction. You can supplement these models with your own experimental pKa data. However, the provided data needs to be prepared using the same format as the training data, that is, the pKa value is assigned to one microtransition and the descriptors are calculated and saved in the same order.
This tutorial uses the Prepare Supporting Experimental pKa Data File Floe. This floe assumes that experimental macro pKas are provided which are associated with a macrotransition (i.e., net charge transitions). This floe uses predicted micro- and macro- pKas to assign the experimental data one dominant microtransition (i.e., a single addition or removal of a proton). The output for this floe will have the descriptors associated with this microtransition associated to the experimental pKa. The output data is saved as an oedb file. If a user would like to see the data, it can be converted to a dataset by the Import File to Dataset Floe.
The output data file can be used as input for the other pKa floes in this package, as described in the Advanced Options parameter sections of the tutorials for the Generate Ionization States and Calculate LogD and pKa Exploration Floes.
The estimated cost of the Prepare Supporting Experimental pKa Data File Floe run is approximately $0.01/molecule.
Running the Floe
Find the Floe
Click the “Floe” button on the navigation bar to reach the Floe page.
Click on the Floes tab.
From the Categories Floe Filters, click on the ‘Packages’ drop-down to expand the list of packages, then select the OpenEye pKa Prediction Floes package.
A list of the pKa Prediction floes will now be visible to the right. Select the Prepare Supporting Experimental pKa Data File Floe.
Alternatively, you can enter the name of the floe in the search bar.

Figure 1. The brief description of the Prepare Supporting Experimental pKa Data File Floe.
Click the “Launch Floe” button to open the Job Form.
Floe Parameters
Specify the parameter settings as indicated below.
Output Path: Select the path where you want to save output of this floe. For this tutorial, use pKa Floes Tutorial/My Data/Tutorial_Output/DataPrep, as shown in Figure 2.
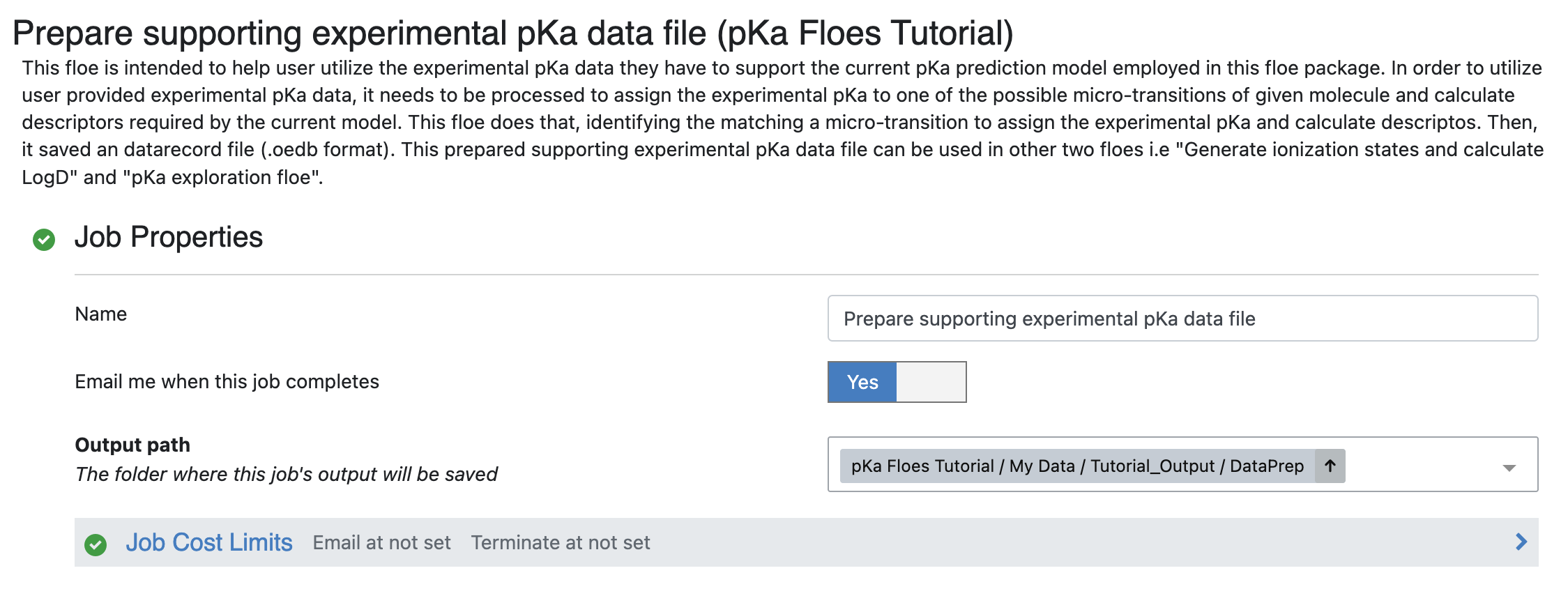
Figure 2. The output path for the floe.
Inputs
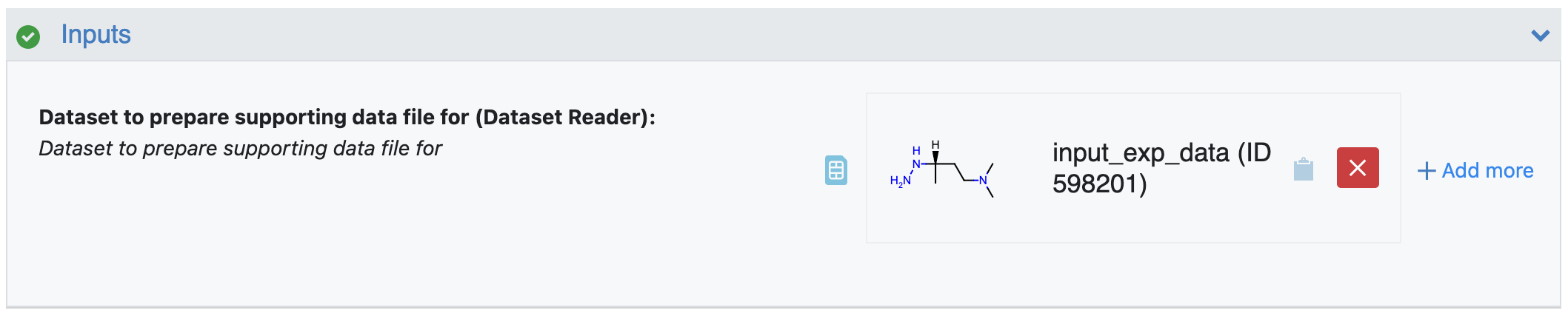
Figure 3. The tutorial input dataset.
Dataset to Prepare Supporting Data File for: This is the input dataset for this floe where the primary molecule will be used to generate corresponding pKa data. The pKa value and its error estimates (if available) should be in specifically titled fields. Those should be in fields named:
pKa and pKa_std (if all records have a single pKa value)
exp_pKas and exp_pKas_std (if single or multiple values are present for a record). Those values should be included as a list of single or multiple values.
For this tutorial, use ‘input_exp_data’ as the input dataset. This sample dataset contains 100 molecules with multiple pKa values in the field named ‘exp_pKas’ and corresponding error values in ‘exp_pKas_std’. Figure 4 shows the molecules in this dataset.
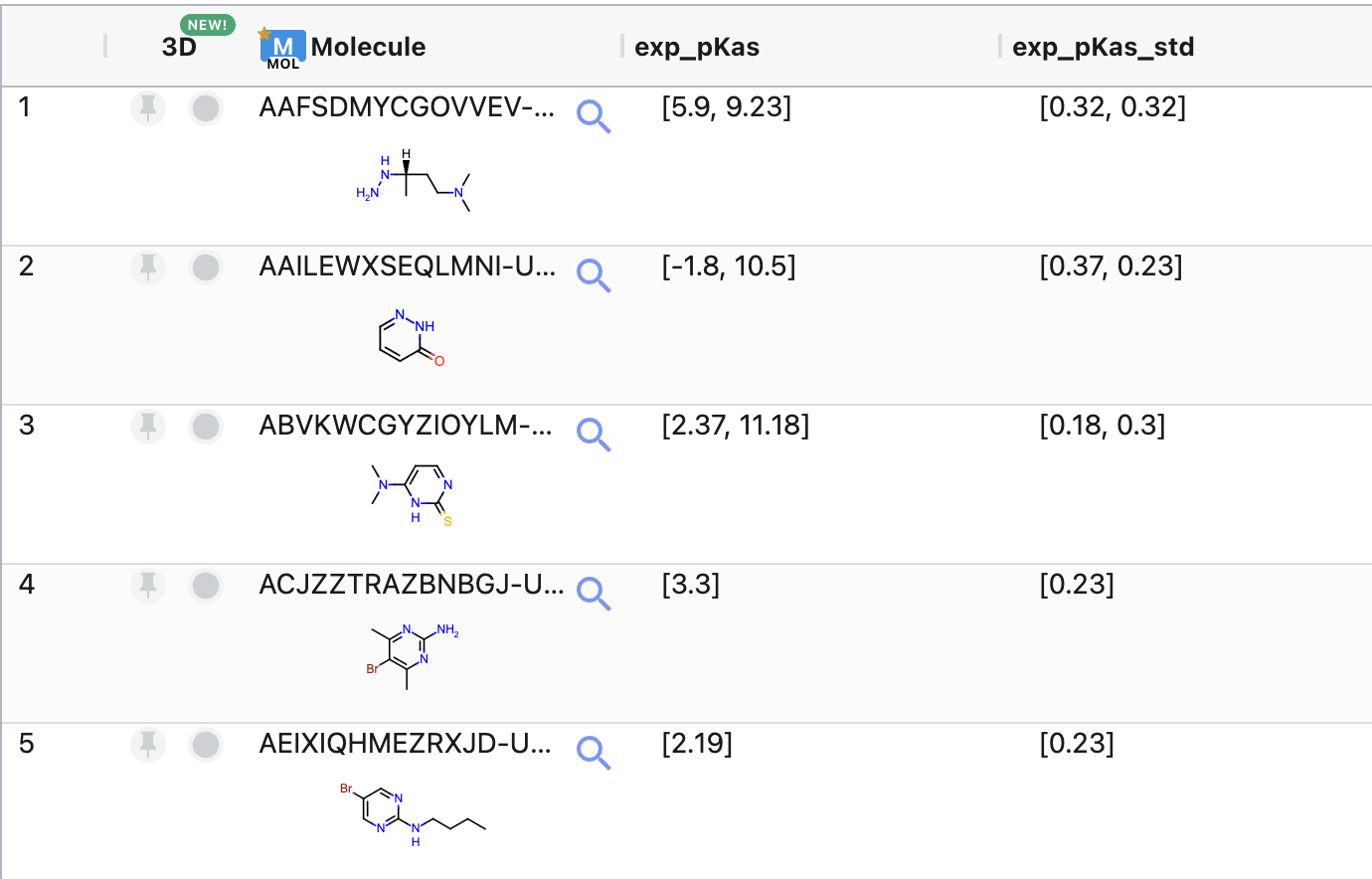
Figure 4. Structures of the molecules in the sample input dataset.
The sample pKa dataset for this tutorial can be downloaded here.
Input Experimental pKa Dataset
Outputs
Specify the names of the output file and failure output datasets here.
Supporting Experimental pKa Data File: Name the prepared output file ‘supporting_exp_data’. There is no need to specify the file extension: the floe will write an oedb file.
Molecule Where Failed to Match Exp Data with Predicted pKa: This parameter assigns an output dataset for instances where the floe failed to match an experimental pKa to a predicted pKa and thus was not able to assign it to any microtransition. Use ‘exp_data_match_failed’ for this tutorial.
Failed descriptors calculation: This parameter assigns an output dataset for instances where the floe fails to calculate descriptors for any molecules. Use ‘exp_data_descriptors_failed’ for this tutorial.
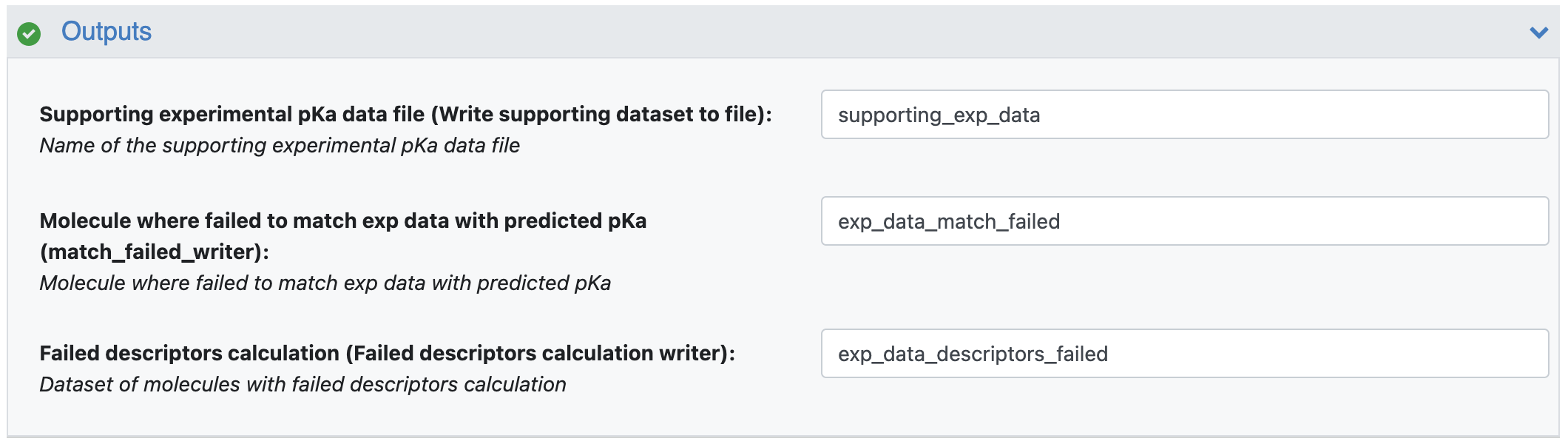
Figure 5. The floe output parameters.
Options
Maximum Difference between Experimental and Predicted pKa: This parameter describes the maximum difference allowed between the experimental pKa and the predicted pKa to match them while preparing dataset. The default value is 1.5. You can modify this value to make matching of the experimental and predicted pKa values to be less or more strict. For this tutorial, keep it at the default value of 1.5.
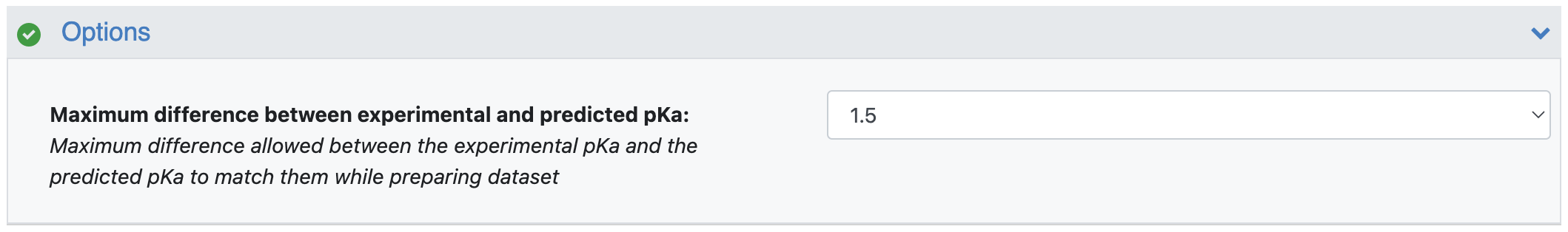
Figure 6. The Options floe parameter.
When all parameters have been set, click the “Start Job” button to run the floe.
Floe Results
Figure 7. An overview of the cubes in the completed floe.
Floe Output Dataset
The floe will have written the prepared data file as supporting_exp_data.oedb. The file was named in the Supporting Experimental pKa Data File parameter under Advanced Options. This is the prepared experimental data file which can be used in the other pKa floes as supporting experimental data to the current model.