GraphemeTM TK 1.3.2
New features
A new function,
OEDrawPeptide, has been added that identifies standard amino acid components in molecules and substitutes them with circular glyphs and corresponding labels. The figure below illustrates this functionality for oxytocin.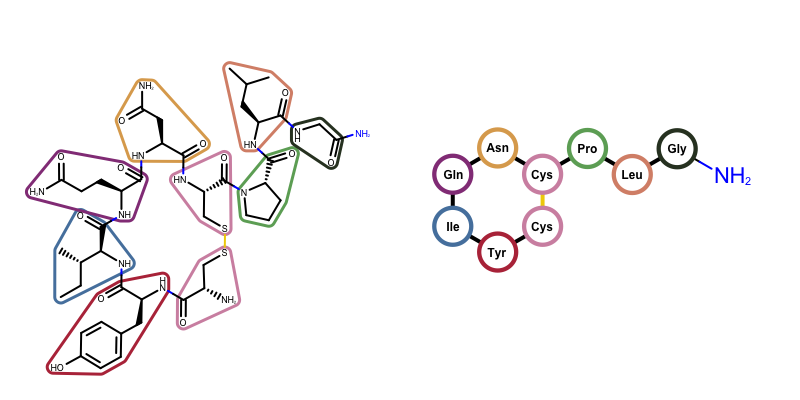
Example of visualizing oxytocin
(left) depiction of oxytocin highlighting its amino acid fragments
(right) depiction of oxytocin using the OEDrawPeptide function
OERenderActiveSitecan now visualize halogen bond interactions (represented by a brown wavy arrow).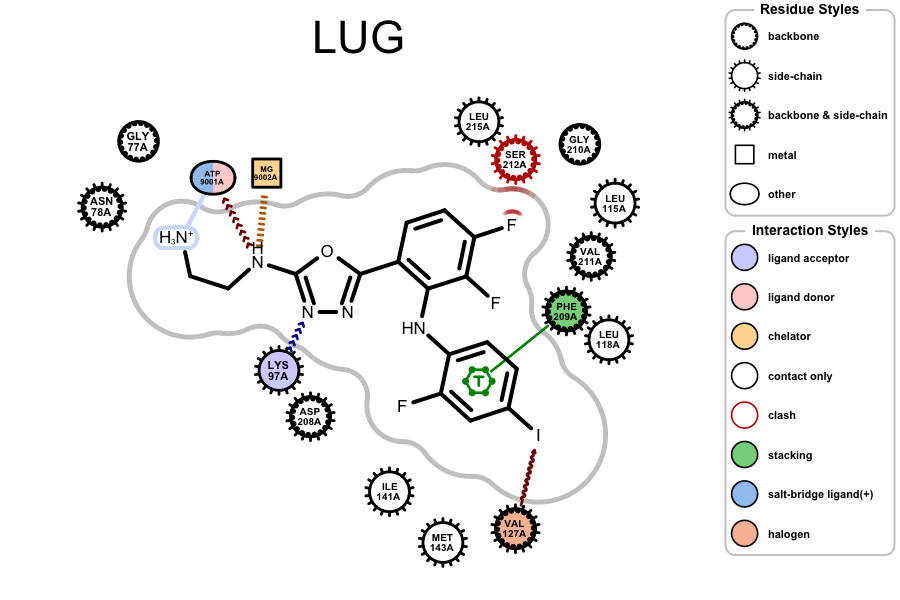
Example of visualizing halogen bond interaction (
PDB 3EQB)The legend of the protein-ligand depiction has been improved by separating and labeling residue styles and interaction styles (see image above).
Minor bug fixes
The label “only contact” has been changed to “contact only” in the legend of the protein-label depiction (see image above).