Searching fast fingerprint database
A program that initializes a fast fingerprint database with the fingerprint file generated from the previous example and then performs a fingerprint search to identify molecules which are similar to a given query molecule.
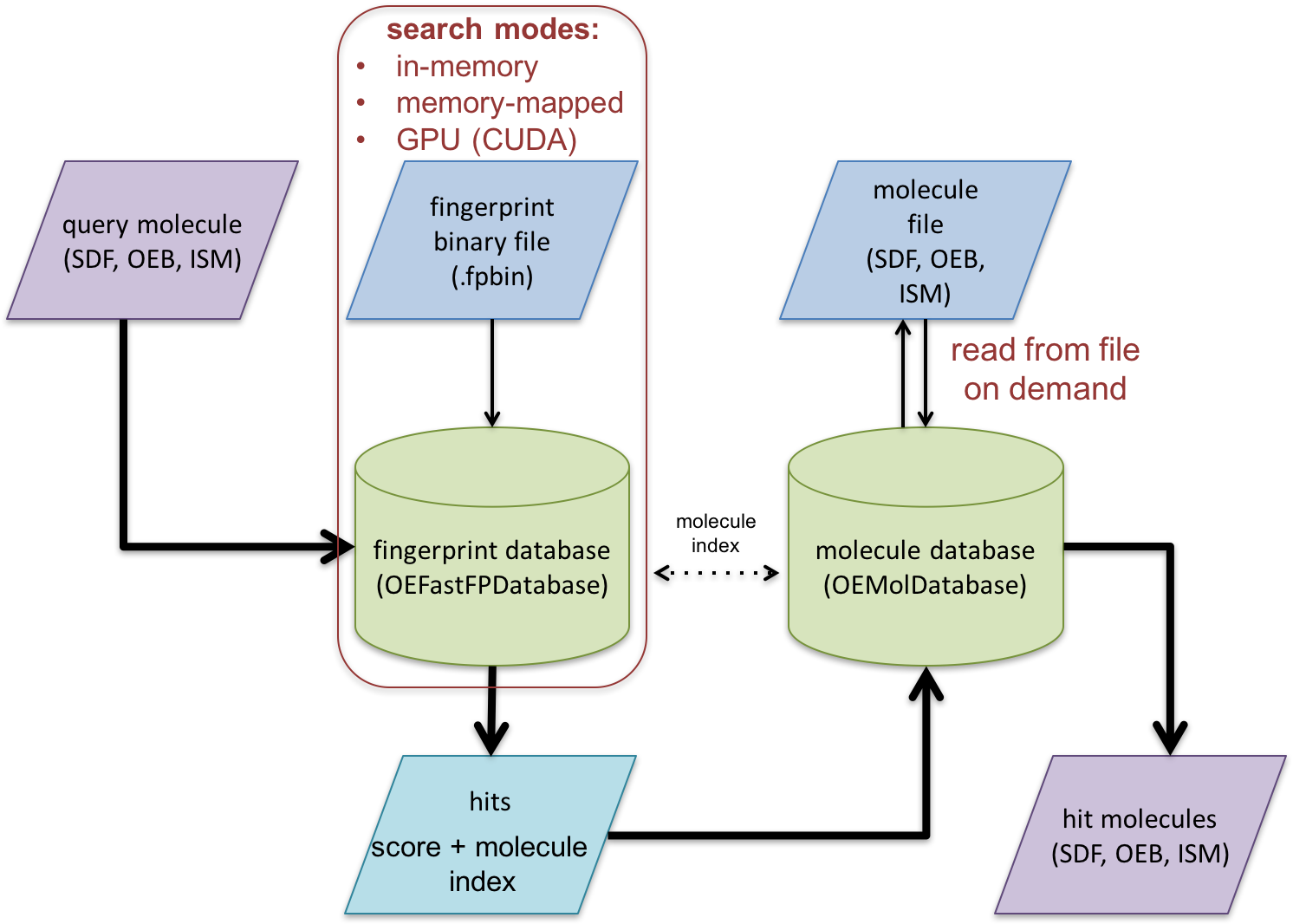
Schematic representation of fast fingerprint search process
See also
Example code in the Generating fingerprint file for fast fingerprint search section
Rapid Similarity Searching of Large Molecule File OpenEye Python Cookbook example
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> java SearchFastFP --help
will generate the following output:
Simple parameter list
fingerprint database options
-memorytype : Fingerprint database memory type
fingerprint database search options
-cutoff : Similarity cutoff value
-descending : Order of similarity scores
-limit : Maximum number of similarity scores
-simfunc : Similarity measure
input/output options
-fpdbfname : Input fast fingerprint database filename
-molfname : Input molecule filename
-out : Output molecule filename
-query : Input query filename
Code
Download code
SearchFastFP.java
and
SearchFastFP.txt
interface file
See also
OEFPDatabaseOptions class
OEConfigureFPDatabaseOptionsandOESetupFPDatabaseOptionsfunctionsOEConfigureFPDatabaseMemoryTypeandOEGetFPDatabaseMemoryTypefunctionOEMolDatabase class in OEChem TK manual
OEFastFPDatabaseMemoryTypenamespaceOEFastFPDatabase class
OEAreCompatibleDatabasesfunction