BROOD Floe Tutorial
The BROOD floe uses BROOD to generate bioisosteric analogs by replacing user-specified portions of the lead with fragments that have similar shape and electrostatics but with potentially novel connectivity and chemistry.
Floes Used in the Tutorials
Input Parameters
The BROOD floe minimally requires a BROOD query and a BROOD fragment database collection. The following section outlines how to prepare a query for the BROOD floe. Users can use the default BROOD fragment database collections available or generate their custom fragment database collection using CHOMP - Generate BROOD Fragment Database. The user can provide an optional input protein for clash check. Any hits with clashes with this protein will be eliminated.
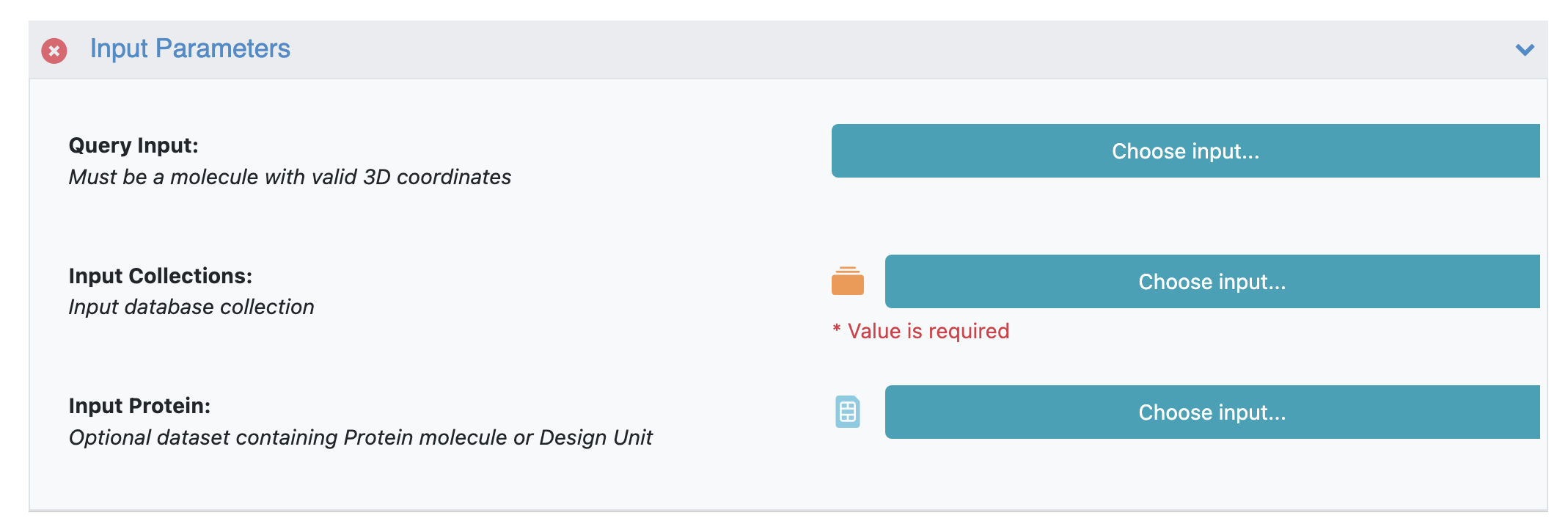
BROOD Floe Input Parameters
BROOD Query Preparation
Clicking on the Choose input prompt for the Query Input parameter will bring up the UI sketcher. Within the UI sketcher, the user can select an existing molecule and make a fragment selection like below.
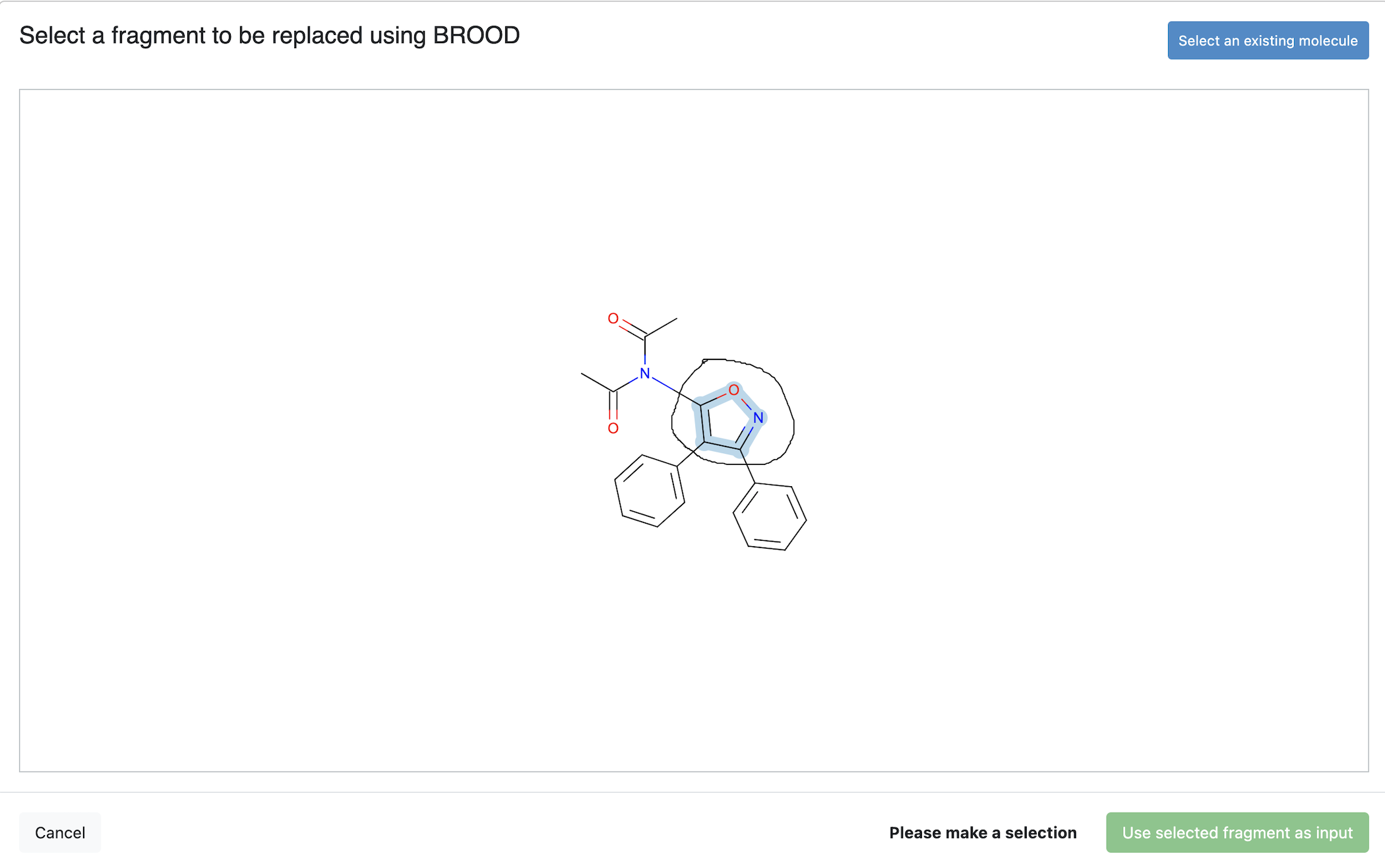
BROOD Query Preparation
The BROOD query can be saved for future runs by turning ON the Save Input Query switch.
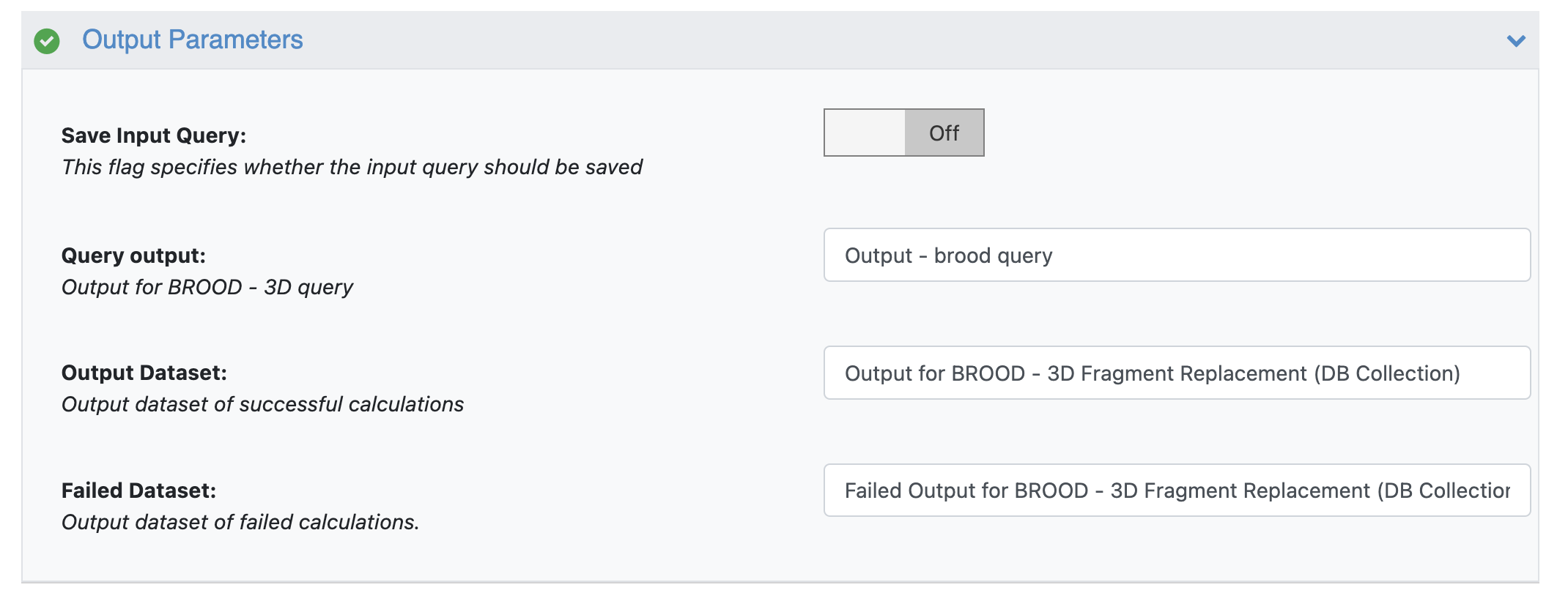
BROOD Floe Output Parameters
Running in ET and linkOnly modes
By default, BROOD generates bioisosteric analogs by finding fragments with similar shape and chemistry. The BROOD floe can be run in ET and linkOnly modes by turning on the appropriate flags.
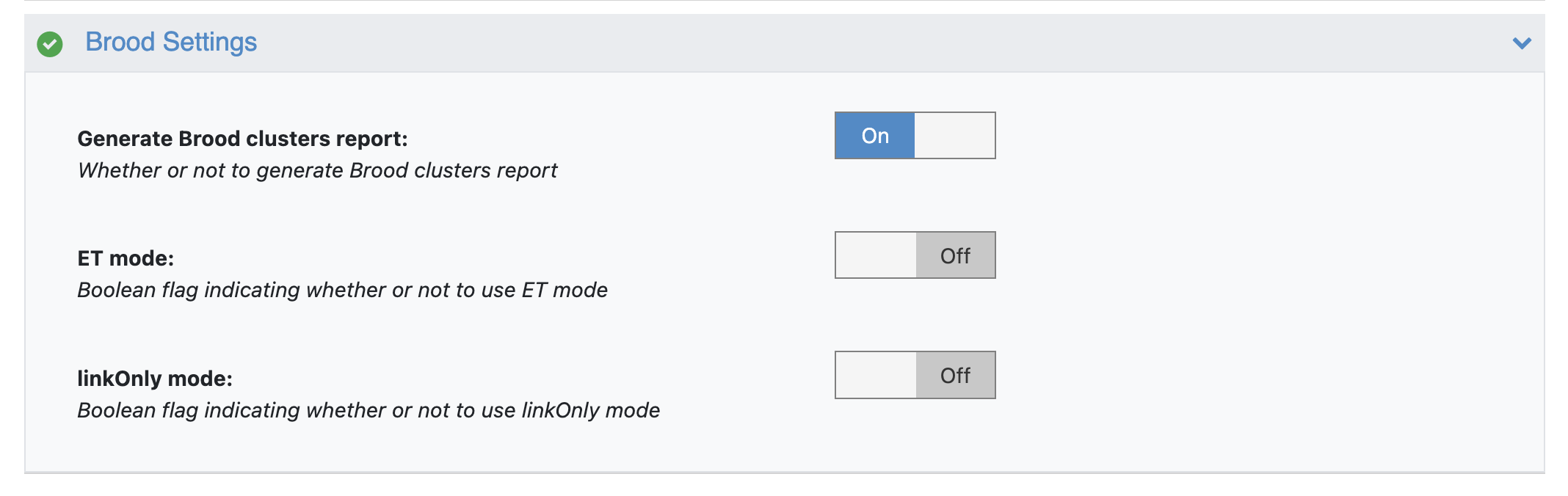
BROOD Floe Settings
Interpretation of Results
The BROOD output hitlist is written to a dataset that can be viewed on the analyze or 3D pages. Additionally, if the Generate Clusters Report flag is turned ON, the output hits are clustered according to a reduced graph hierarchy. The cluster heads representing the best-scoring member of each cluster can be viewed in the floe report.
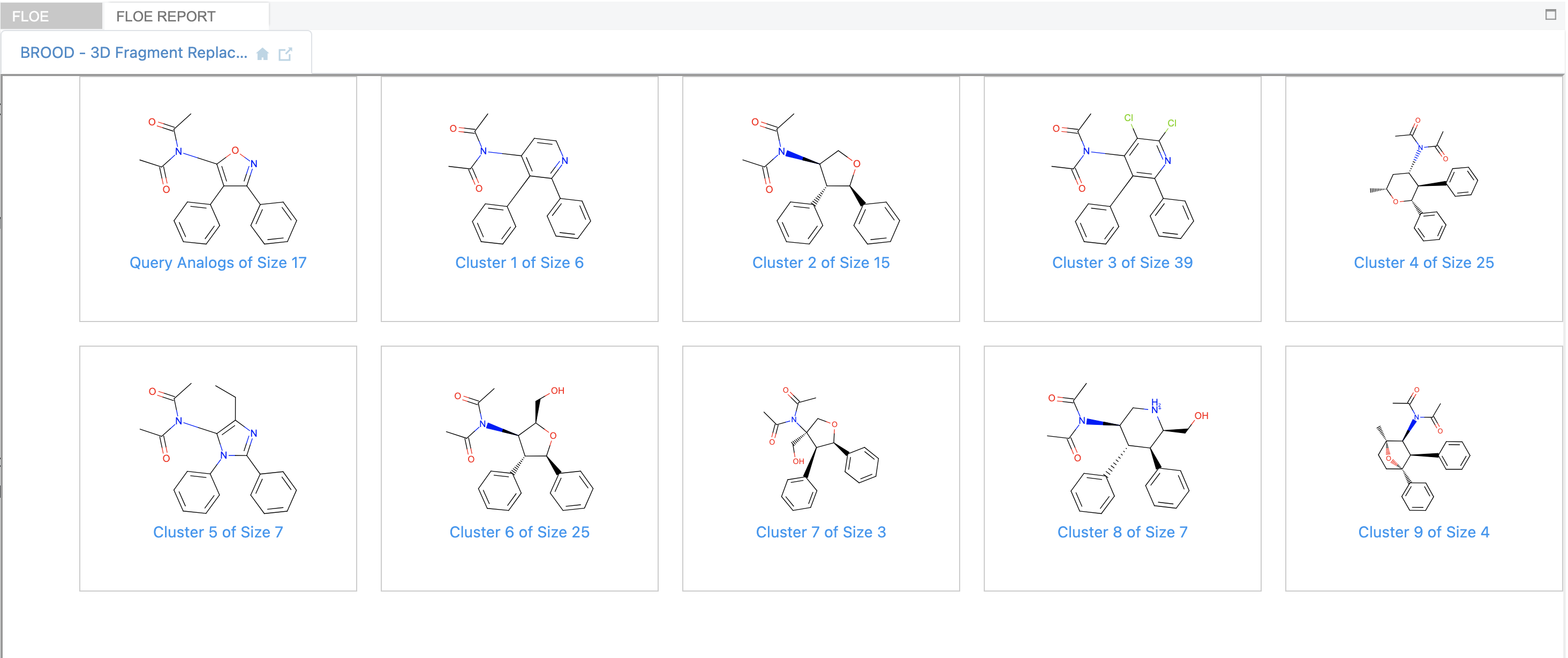
BROOD Floe Report