Prepare Vendor Database for Giga Docking and FastROCS
In this tutorial compounds from the Mcule Purchasable (In Stock) database will be prepared for Giga Docking and FastROCS. Running the floes in this tutorial will generate approximately $45 in Orion compute charges. (the cost will vary somewhat depending on current AWS pricing).
This tutorial uses the following floes
Prepare Giga Collections from the Openeye-large-scale-floes package
Collection Info from the OpenEye-large-scale-floes package
Sample Collection from the OpenEye-large-scale-floes package
Note
OpenEye already prepares several vendor collection for use in Orion (as of this writing Enamine Real, Wuxi Galaxy and Mcule Ultimate). This tutorial is for those who wish to prepare their own custom in-house databases or other vendor database for Giga-Docking and fastROCS.
Create a Tutorial Project and Working Directory
Note
If you have already created a Tutorial project while doing another tutorial you can re-use the existing one and skip this step.
Log into Orion
Click the Home button at the top of the left menubar.
Click on the ‘Create New Project’ button and in the pop up dialog enter Tutorial for the name of the project and click ‘Save’.
Import Mcule Database into Orion
Download the Mcule Purchasable (In Stock) database in .smi.gz format from the Mcule database website to your local machine. Then upload the file from your local machine to Orion as follows
Click on the ‘Data’ button in the left menu bar
Select ‘My Data’ from the list of options to the left of the page.
Create an ‘Input Data’ folder if one does not already exist by clicking the blue folder icon with the + symbol.
Open the ‘Input Data’ folder by clicking on it.
Set the ‘Type’ drop down menu to Files if it is not already.
Open the ‘Add Data’ menu and click ‘Upload’
In the modal that appear select the Mcule Purchasable (In Stock) database file on your local machine.
Select ‘Show advanced options’
Select ‘None’ for the processing method of the file.
Warning
If ‘None’ is not selected here the uploading of the file may fail.
Click ‘Upload’
Important
As of the writing of this document the Mcule Purchasable (In Stock) database is ~10 million molecules, and preparing these molecules will cost approximately $50 in Orion compute charges. If at the time you download this database it is larger than ~10 million the Orion compute charges will be higher (the cost is roughly linear with the number of molecules).
Create GigaDocking and FastROCS collections
Locate the Prepare Giga Collections floe as follows
Click on the ‘Floes’ button in the left menu bar
Click on the ‘Floes’ tab in the upper left of the main window.
Click ‘All Floes’ in the left pane
In the search bar at the top of the right pane enter Prepare Giga Collections
The Prepare Giga Collections will be visible to the right. Click it and enter the following parameter settings in the floe launch UI that pops up.
Jobs Properties
Output Folder : Tutorial/My Data/Mcule In Stock Collections
Promoted Parameters
Inputs
Input File(s) (Semi-Optional) : Tutorial/My Data/Input Data/mcule_purchasable_in_stock_210615.smi.gz
To select this file you downloaded from the Mcule site and then upload to orion.
Note
The file you downloaded from Mcule may have a slightly different name than mcule_purchasable_in_stock_210615.smi.gz.
Click the ‘Choose Input Button’
Click on ‘Project Data’ under collections in the Select Input File(s) modal.
If you do not see the mcule_purchasable_in_stock_201103.smi.gz then in the search bar type ‘mcule’ and click ‘search for resource mcule’ in the drop-down menu.
Select the mcule_purchasable_in_stock_201103.smi.gz file.
Click ‘Use File as Input’
Outputs
Giga Docking Collection Name : Mcule In Stock GigaDock Collection
FastROCS Collection Name : Mcule In Stock FastROCS Collection
Now launch the floe using the ‘Start Job’ button at the bottom of the floe launch UI. This job may take up to 10h to complete and will have an Orion Compute Charge cost of approximately $40.
The output of this floe will be the following two collections:
Mcule In Stock GigaDock Collection : Molecule ready for use with the Giga Docking floe.
Mcule In Stock Purchasable FastROCS Collection : Molecule ready for use with the
See also
Note
If you are using this tutorial as a guide to prepare your own set of molecules and the format of the input files is CSV see Prepare Molecules in a CSV File for GigaDocking and FastROCS in addition to this tutorial.
View Collection Properties
Collections cannot be introspected by the Orion UI like dataset can. To view basic information (e.g., Molecular Properties) of the prepared collections first located the Collection Info floe
Click on the ‘Floes’ button in the left menu bar
Click on the ‘Floes’ tab in the upper left of the main window.
Click ‘All Floes’ in the left pane
In the search bar at the top of the right pane enter Collection Info
The Collection Info will be visible to the right. Click it and enter the following parameter settings in the floe launch UI that pops up.
Jobs Properties
Output Folder : Tutorial/My Data/Mcule In Stock Collections
Promoted Parameters
Inputs
Input Collection : Tutorial/My Data/Mcule Purchasable Collections/Mcule Purchasable In Stock GigaDock Collection
Select the collection generated in the previous step of the tutorial as follows.
Click the ‘Choose Input’ button for Input Collection to open the Select Collection modal.
Select the ‘My Data’ folder.
Select the ‘Mcule Purchasable Collections’ folder.
Select the Mcule Purchasable In Stock GigaDock Collection collection.
Click ‘Use dataset as Input’.
Note
You could also select the ‘Mcule In Stock FastROCS Collection’ instead of the GigaDock collection. It will have nearly identical molecular properties to the Gigadock collection, but will have a maximum of 10 conformers per molecule rather than 200.
The floe will take about 15min to run and with an Orion compute charge of about $1. Once the floe completes go to the job page and click on the ‘Floe Report’ tab above the window with the floe diagram. A set of histograms showing the basic molecular properties of the collection will then be visible.
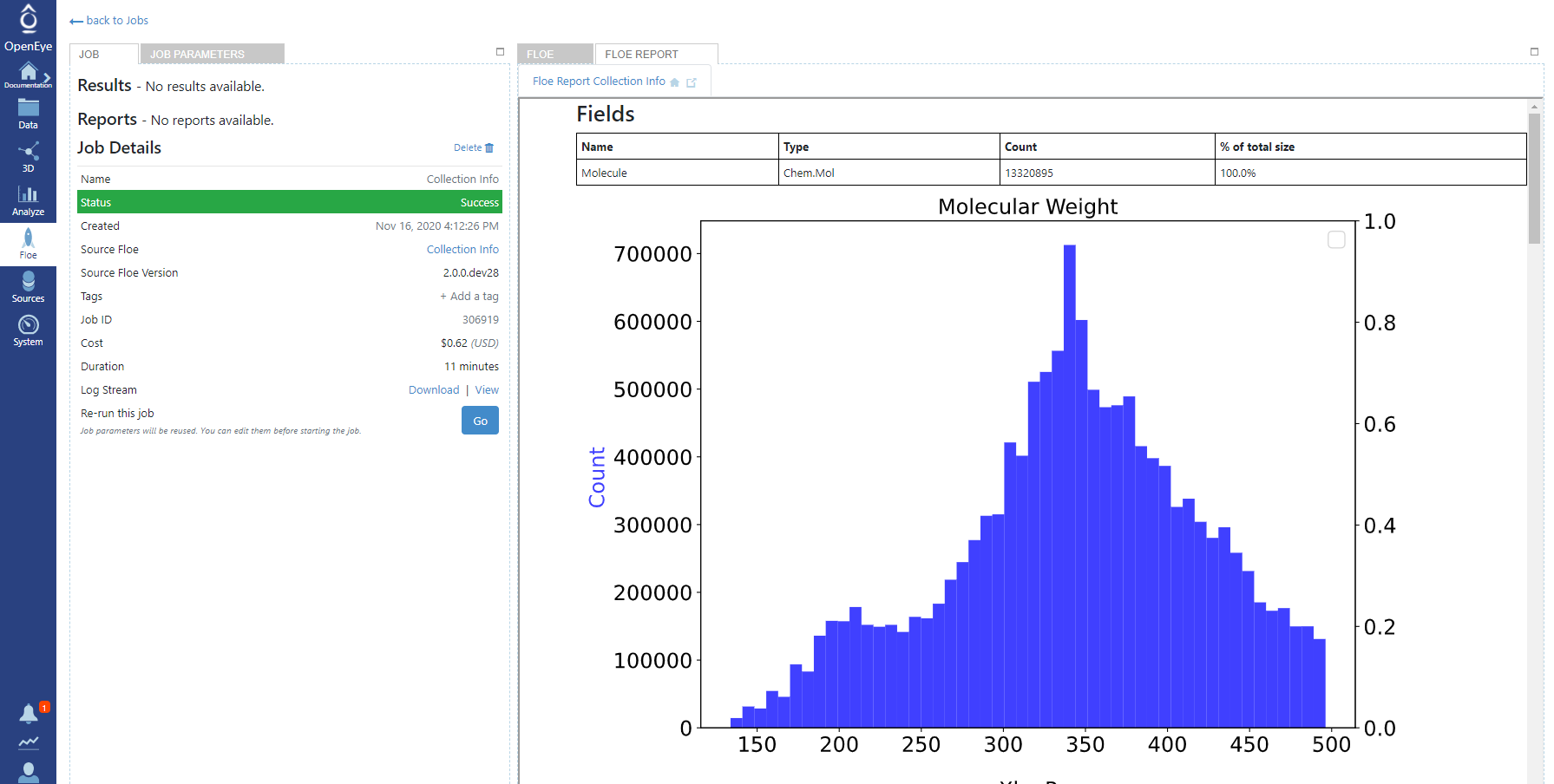
Image of the floe report on the Mcule collection.
View of Random Sample of the Collection
The previous step in the tutorial examining the distribution of basic molecular properties of the prepared molecules. In this step a random sample of the collection will be converted into a dataset and examined in the 3D page.
Note
Collection are not viewable in the 3D or Analyse page directly.
Located the Sample Collection as follows
Click on the ‘Floes’ button in the left menu bar
Click on the ‘Floes’ tab in the upper left of the main window.
Set the ‘Browse Workfloes’ drop down menu to ‘Show all packages’
Select ‘All’ under Browse Workfloes
In the search bar enter Sample Collection
The Sample Collection will be visible to the right. Click it and enter the following parameter settings in the floe launch UI that pops up.
Jobs Properties
Output Folder : Tutorial/My Data/Mcule In Stock Collections
Promoted Parameters
Inputs
Input Collection : Tutorial/My Data/Mcule Purchasable Collections/Mcule Purchasable In Stock FastROCS Collection
Select the collection generated in the previous step of the tutorial as follows.
Click the ‘Choose Input’ button for Input Collection to open the Select Collection modal.
Select the ‘My Data’ folder.
Select the ‘Mcule Purchasable Collections’ folder.
Select the Mcule Purchasable In Stock FastROCS Collection collection.
Click ‘Use dataset as Input’.
Note
You could also select the ‘Mcule In Stock GigaDock Collection’ instead of the FastROCS collection. It will have nearly identical molecular properties to the FastROCS collection, but will have a maximum of 200 conformers per molecule rather than 10.
The floe will take about 15min to run and with an Orion compute charge of about $1. Once the job finishes view the sample in the 3D view as follows
Navigate to the floe job page
Under Results in the top left click the ‘Show in Project Data’ link next to ‘Sample Dataset’
Clear any currently active datasets by opening the ‘Active Datasets’ drop down un the upper left and clicking ‘Clear All’
Make the Sample Dataset active by clicking on the + symbol to the left of the name to make it green.
Switch to the 3D view by clicking the ‘3D’ button on the blue menu to the left of the interface.
Open the layout menu in the upper right and select ‘3D viewer with spreadsheet’
You can now browse individual records using the mouse or up down arrows.