CHOMP Floe Tutorial
The CHOMP floe uses CHOMP to fragment molecules and generate a BROOD fragment database collection, which can be used as an input in BROOD - 3D Fragment Replacement.
Floes Used in the Tutorials
Input and Output Parameters
The CHOMP floe requires an input of 2D molecules in a dataset, a file, or a collection.
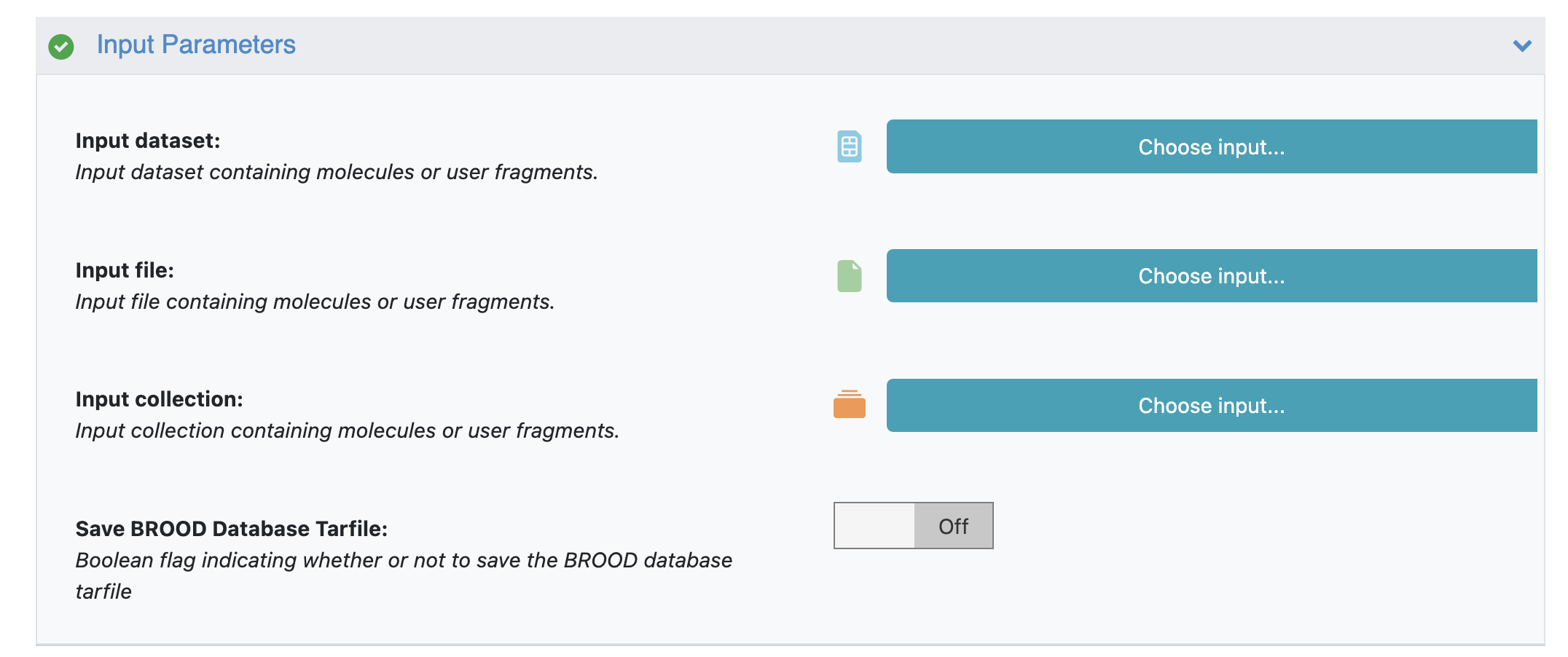
CHOMP Floe Input Parameters
The output of the CHOMP floe is a BROOD fragment database collection. There’s also an option to save the BROOD fragment database tar file by turning on the Save BROOD Database Tarfile switch. Please ensure that the Output database name does not contain spaces; otherwise, the floe will fail. This output tar file can be downloaded, extracted, and used locally as input for the BROOD application.
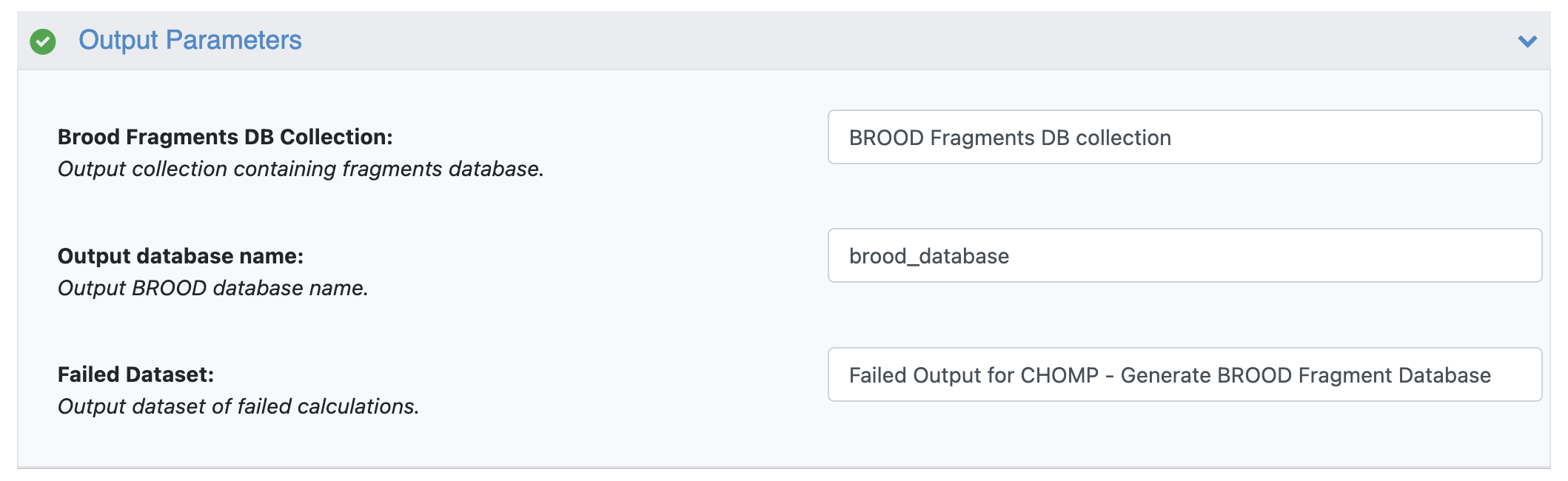
CHOMP Floe Output Parameters
Fragment Filtering Parameters
The fragment filtering options decide what fragments are retained in the final BROOD fragment database. By default, the CHOMP floe has been set up to create the lite version of the BROOD fragment database. In more detail, all fragments containing 9 heavy atoms or less, and fragments having more than 9 heavy atoms with a frequency of at least 3 are retained. These settings can be changed by editing the fragment filtering parameters. Turning on the Use Percentile switch filters the fragments by treating the minimum frequency input as a percentile.
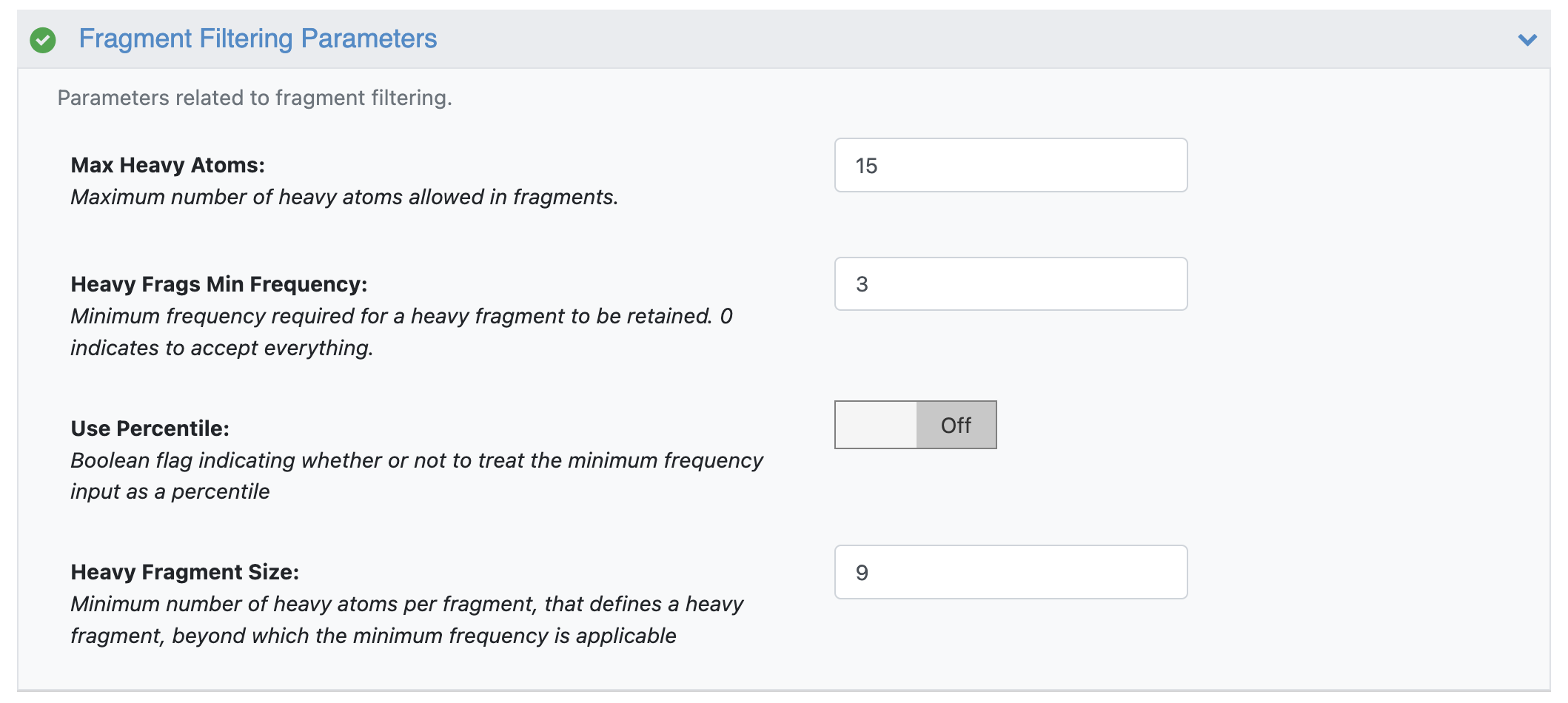
CHOMP Floe Fragment Filtering Parameters
Guidelines for Control Parameters
The default values for control parameters have been set up to handle ~1 million drug-like molecules. When the input has more than ~1 million drug-like molecules, set the control parameters following the guidelines below.
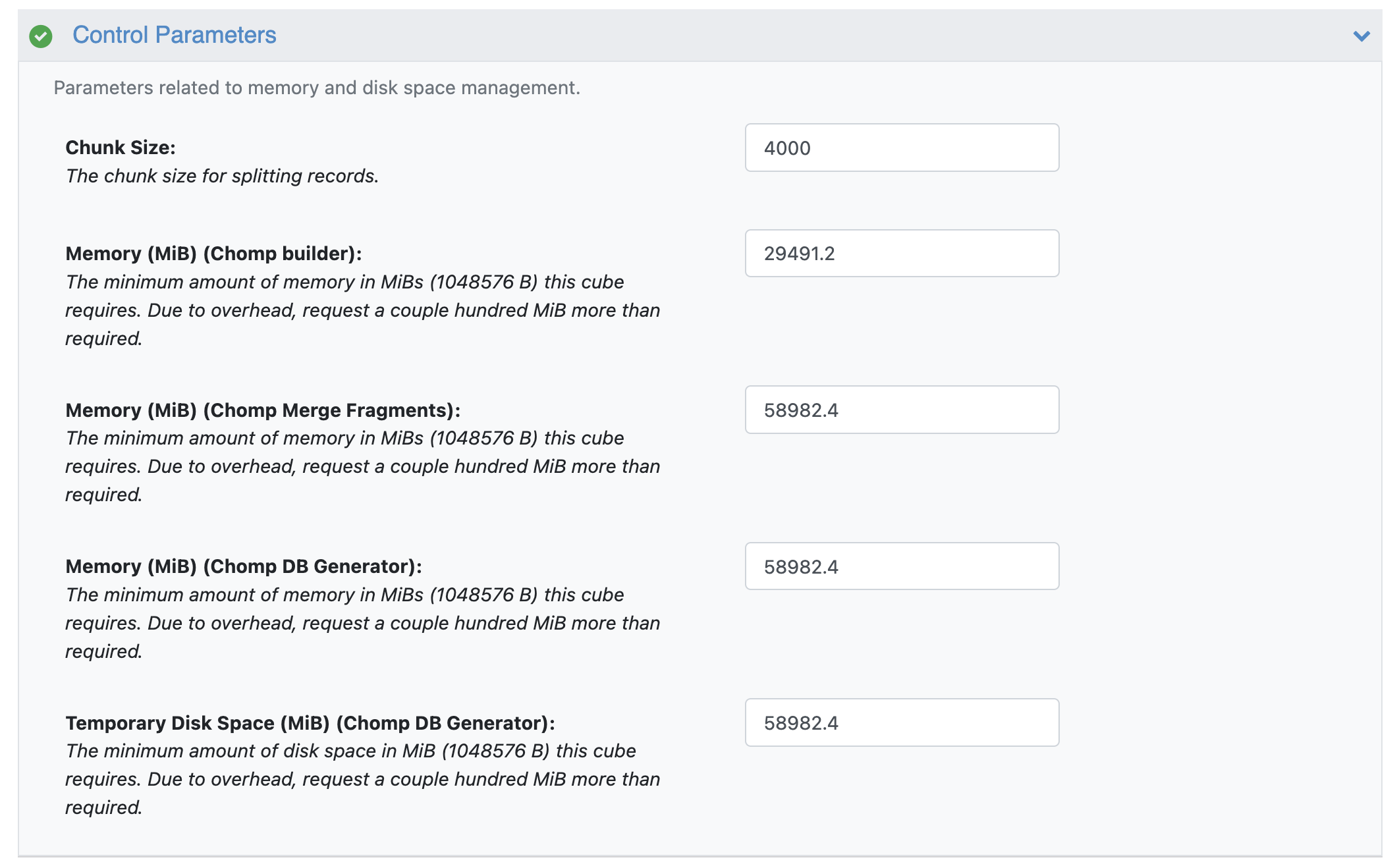
CHOMP Floe Control Parameters
Chunk Size: Set the chunk size so the number of chunks is ~250. For example, for ~1 million drug-like molecules, the suggested chunk size is the default value of 4000.
Memory (MiB) (Chomp builder): Multiply the default memory value by the ratio of change in chunk size. For example, if the default chunk size is doubled, multiply the default memory by 2.
Memory (MiB) (Chomp Merge Fragments): Set this value to ~0.01 times the number of fragments.
Memory (MiB) (Chomp DB Generator): Set this value to ~0.009 times the number of fragments.
Temporary Disk Space (MiB) (Chomp DB Generator): Set this value to ~0.009 times the number of fragments.
Please note that these guidelines are approximate, and the specific values may differ for each input. A recommended 10% increase in memory and disk space values is advised to provide a margin of safety.
Interpretation of Results
The BROOD Fragment Database Collection is the most important output of the CHOMP floe, containing the final 3D conformers of the fragments. The Brood Fragment Database Report reports the final number of fragments in the BROOD Fragment Database Collection. The failed outputs are written to a dataset and detailed in a failure report.