Large Scale Clustering
The large scale clustering floes provide a relatively efficient method to cluster for large datasets. This tutorial will discuss how to successfully cluster a large dataset using the large scale floes, focusing on the input parameter settings that can be optimized.
Floes used in this Tutorial
The floes used in this tutorial are:
Input Size
The large scale floes are designed for input datasets with more than 20,000 molecules, and in most cases will complete with several hundred thousand molecules. For 3D input, only a single conformer is considered, so keep that in mind for dataset size. If your input dataset is smaller than 20,000 molecules, it is recommended to run another one of the clustering floes. To assist your decision, see the clustering decision tutorial.
Algorithm: Directed Sphere Exclusion
The algorithm used in large scale clustering is called directed sphere exclusion [1]. It involves two steps that repeat in a cycle:
Step 1: Generate N new cluster heads
Step 1 is carried out In a serial cube. A set number N of cluster head molecules, determined by the cluster head ratio parameter and the batch size parameter, is found by this cube.
The molecule on the first input record is chosen as the first cluster head. From there, new cluster heads are found by comparing this cluster head, and any further ones found, to unassigned molecules. If a molecule is within the sphere exclusion radius of a cluster head, it is assigned to that cluster head, and sent out of the cycle. Otherwise, it is added to the cluster head list. Once a set number of heads has been found, the remaining unassigned members are sent out of the cube, to Step 2.
This step 1 also involves sorting the list of remaining molecules before looking for new cluster heads. If an optional score field is selected, before head selection, the molecules are sorted using this score field. Otherwise, molecules are sorted by their similarity to the molecule on the arbitrary first input record.
Step 2: Assign cluster members to existing cluster heads
Step 2 is carried out in instances of a parallel cube. Each instance of the cube has a list of all the cluster heads found so far, and a set number of unassigned molecules, determined by the item_count parameter. These heads are compared in order to unassigned molecules, and if a molecule is within the sphere exclusion radius of a cluster head, it is assigned to that cluster head, and sent out of the cycle.
If a molecule is not assigned to a cluster head, it continues within the cycle to step 1. Clustering completes when there are no longer unassigned molecules, or in rare cases, to prevent indefinitely long runs, when the cycle has been repeated more than 10,000 times.
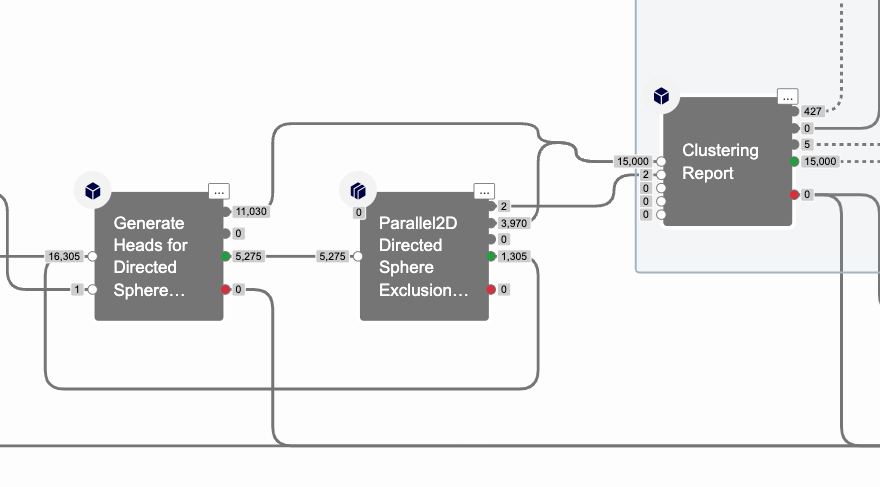
Sphere Exclusion Cube Cycle
Sphere Exclusion Radius
The sphere exclusion radius is the most important input parameter to consider in any of the large scale floes. It is compared to the distance between two molecules, which is 1.0 - <the similarity score of the molecules> . In step 2 above, if an unassigned molecule is within the radius of an existing cluster head, it is assigned to that molecule. If not, it is compared to other cluster heads (in step 2 above) and if it remains unassigned (outside of the sphere exclusion radii of all existing cluster heads), it is sent back to head generation may be chosen as a new cluster head (in step 1 above).
A lower sphere exclusion radius will result in a greater number of clusters, each with fewer members. A higher radius will result in fewer clusters, each with more members. It may take some initial runs of this Floe to determine the appropriate radius for your input dataset.
Troubleshooting
This section suggests some solutions for potential problems users may encounter running a large scale clustering floe.
Floe Runtime is Too High
This problem is most likely caused by choosing a sphere exclusion radius that is too low or too high.
If the sphere exclusion radius is too low, then the generate heads cube will run much longer for each batch it processes, as it will take much longer to find the number of cluster heads. Other than increasing the radius, one potential remedy for this situation is to decrease the cluster head percentage and/or the minimum number of cluster heads.
If the sphere exclusion radius is too high, there will be a large number of cluster heads, which will increase memory demand, and reduce the speed of both the parallel sphere exclusion cube in step 2 and the generate heads cube in step 1. The floe may eventually exceed memory limits. For exceptionally large datasets, it may be useful to filter the dataset first, or run the diverse subset floe to find a representative dataset for clustering.
Floe Cost is Too High
This problem is most likely caused by increasing the advanced memory parameters above their default setting of 8000. Instances of cubes with higher memory requirements will be more costly. If the memory needs to be this high, it may also help reduce the cost by attempting to reduce the floe runtime.
Memory Limit Exceeded
This problem is typically caused by more than 10,000 cluster heads found while the floe is running. There are two options to deal with this problem. The first is to increase the sphere exclusion radius, which will decrease the number of cluster heads. The second is to increase the limit for both advanced memory parameters.
Advanced Parameter Descriptions
These advanced parameters may be useful in situations where the floe has problems. See the troubleshooting section section for recommendations.
Batch Size: Batch size is the initial number of records that the generate heads cube will process at a time, to look for cluster heads. This is adjusted automatically based on batch size percentage, once all input records have been counted.
Memory (MB) for Cluster Head Generation: This is the memory available for the process described in step 1. It should be at least as high as the next memory parameter. It may need to be increased in the case of more than 10,000 cluster heads, which happens when there is both a large input dataset and a relatively low sphere exclusion radius.
Memory (MB) for Parallel Sphere Exclusion: This is the memory available for the process described in step 2. It should be at least as high as the previous parameter.
Sphere Exclusion Item Count: This is the number of records that each instance of the sphere exclusion cube, described in Step 2, receives. Each record corresponds to comparing the similarity of one unassigned member to all clusters heads found so far. For a large number (1000+) of cluster heads, it may need to be decreased, but typically it should not be modified.
Cluster Head Percentage: The generate heads cube, described in step 1, looks through batch size records at a time. It stops looking for cluster heads in a given batch when a number of cluster heads equal to this number times the batch size has been found.
Minimum Batch Size: As the floe runs, the batch size is automatically adjusted to be smaller, as the number of records to be processed decreases. This parameter determines the minimum number of records in a given batch.
Minimum Cluster Heads Per Cycle: As the floe runs, the number of cluster heads found per cycle is automatically adjusted to be smaller, as the number of records to be processed decreases. This parameter determines the minimum number of cluster heads to be found in a given batch.
Batch Size Percentage: After the floe counts the number of input records, this parameter determines the batch size as a percentage of that number.
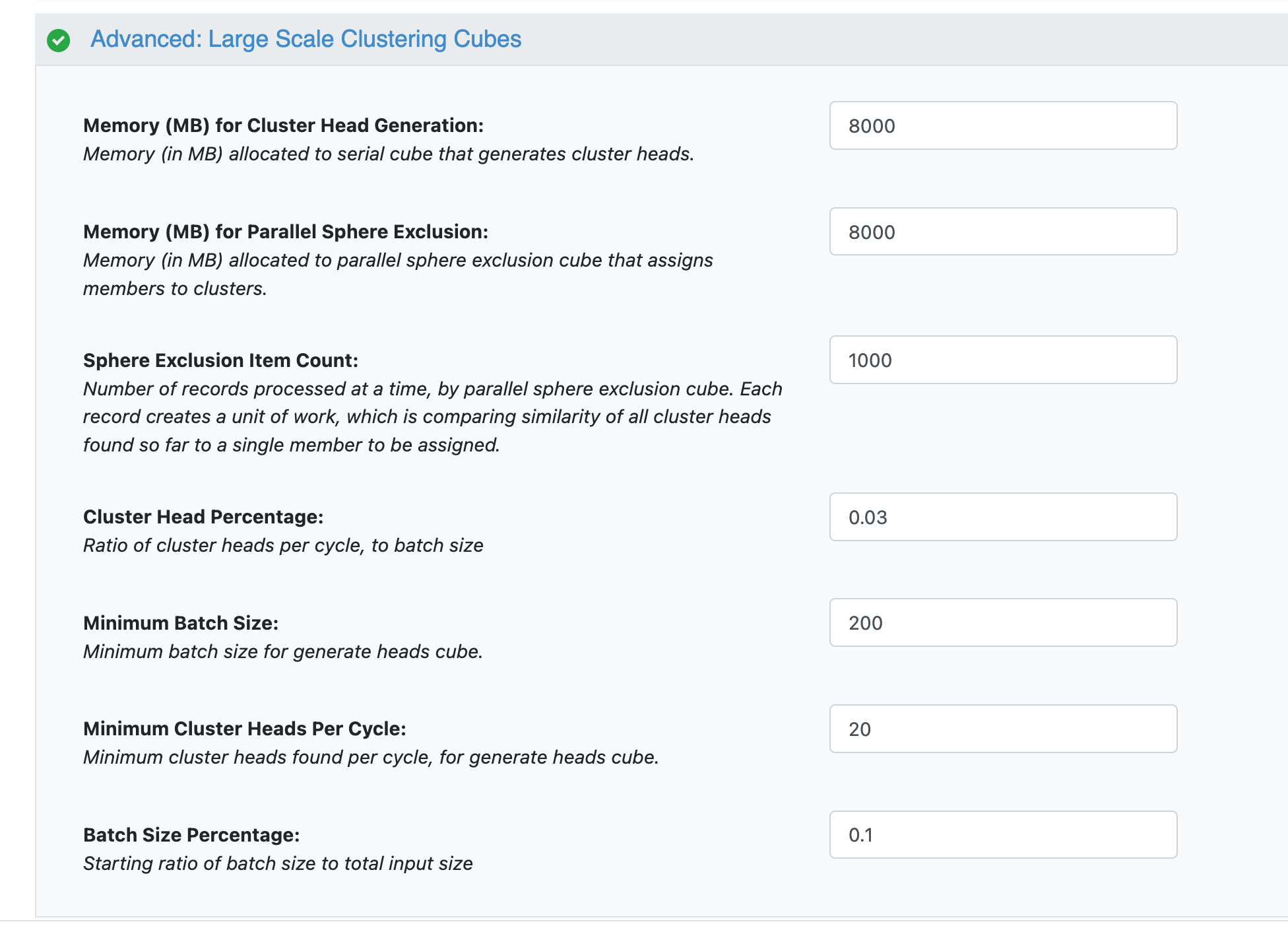
Advanced Clustering Parameters
References
1. DISE: Directed Sphere Exclusion Alberto Gobbi and Man-Ling Lee Journal of Chemical Information and Computer Sciences 2003 43 (1), 317-323 DOI: 10.1021/ci025554v