Determine if Giga Docking Will Give Good Results with a Given Receptor
Context
Docking, like all virtual screening tools, can work better on some systems than others. Before a full Gigadock run is performed the performance of the run can be estimated if there are molecules known to be active against the target available.
Note
If there are several receptors available for the given target (e.g., receptors with different constraints set or receptors using different conformations of the protein) it is recommended that this procedure be used on each receptor, and the receptor with the best enrichment be used for the full run.
Procedure
Use the Filter Collection floe to create a collection of ~100,000 molecules randomly selected from the Giga Docking collection to be used in the full Giga Docking run. See the Dock One Million Molecules with Gigadock Floe tutorial for an example of how this is done.
Generate conformers of the known active molecules using the Classic Omega floe using the default ‘Classic’ conformer generation mode.
Run the Gigadock floe with the following parameters set.
Job Properties
Output Path : Create a new appropriate folder.
Promoted Parameters
Inputs
Receptor Dataset : Select the dataset with receptor.
Input Conformer Collection : Set this to the collection created in step #1.
Actives (Optional) : Set this to the dataset of active conformers created in step #2.
Once the Gigadock floe finishes an ‘AUC’ dataset will be created which contains the AUC value for how well the active molecules score compared to the 100K collection of molecules (obviously some of the 100K may be active, but it is reasonable to assume that the vast majority are inactive and treat it as a background decoy set).
The floe report for the Gigadock job will also contain a histogram showing visually how the active molecules scored compared to the background 100K collection of molecules.
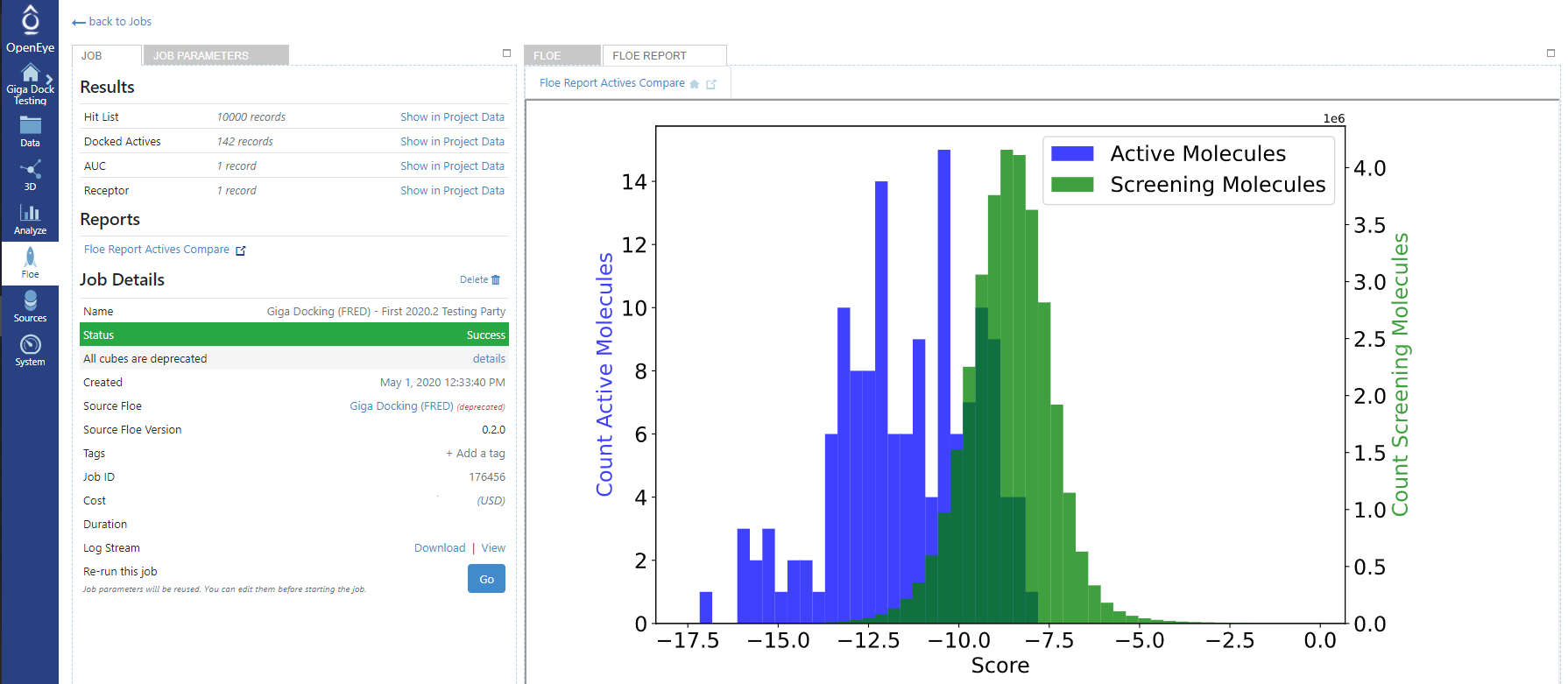
Example floe report