Depicting a Single Molecule
A program that converts a molecular structure into an image file.
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> Mol2Img --help
will generate the following output:
Simple parameter list
image options :
-height : Height of output image
-width : Width of output image
input/output options :
-in : Input filename
-out : Output filename
-ringdict : User-defined 2D ring dictionary
molecule display options :
-aromstyle : Aromatic ring display style
-atomcolor : Atom coloring style
-atomlabelfontscale : Atom label font scale
-atomprop : Atom property display
-atomstereostyle : Atom stereo display style
-bondcolor : Bond coloring style
-bondprop : Bond property display
-bondstereostyle : Bond stereo display style
-hydrstyle : Hydrogen display style
-linewidth : Default bond line width
-protgroupdisp : Protective group display style
-scale : Scaling of the depicted molecule
-superdisp : Super atom display style
-titleloc : Location of the molecule title
prepare depiction options :
-clearcoords : Clear and regenerate 2D coordinates of molecule(s)
-orientation : Set the preferred orientation of 2D coordinates
-suppressH : Suppress explicit hydrogens of molecule(s)
Code
Download code
See also
Molecule Depiction chapter
OEConfigureImageOptionsfunctionOEConfigurePrepareDepictionOptionsfunctionOEConfigure2DMolDisplayOptionsfunctionOEIsRegisteredImageFilefunctionOESetupPrepareDepictionOptionsfunctionOEPrepareDepictionfunctionOEGetImageWidthandOEGetImageHeightfunctionsOE2DMolDisplayOptions class
OESetup2DMolDisplayOptionsfunctionOE2DMolDisplay class
OERenderMoleculefunction
Examples
prompt> Mol2img -in nexium.ism -out image.png
will generate the image shown in Figure: Example of using the program with default parameters.
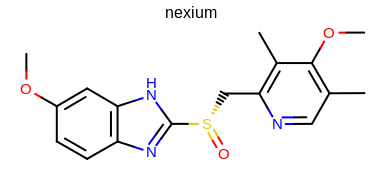
Example of using the program with default parameters
prompt> Mol2Img -in nexium.ism -out image.png -atomcolor WhiteMonochrome
will generate the image shown in Figure: Example of using the program with the white monochrome atom coloring style.
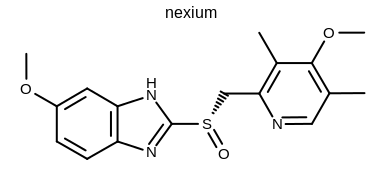
Example of using the program with the white monochrome atom coloring style
prompt> Mol2Img -in nexium.ism -out image.png -aromstyle Circle
will generate the image shown in Figure: Example of using the program with aromaticity style ‘Circle’.
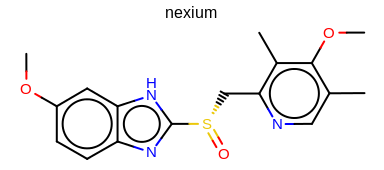
Example of using the program with aromaticity style ‘Circle’
prompt> Mol2Img -in nexium.ism -out image.png -atomstereostyle "AtomStereo|CIPAtomStereo"
will generate the image shown in Figure: Example of the mol2img program with CIP atom stereo option.
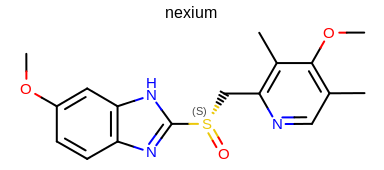
Example of using the program with CIP atom stereo option
prompt> Mol2Img -in nexium.ism -out image.png -linewidth 4
will generate the image shown in Figure: Example of using the program with user-defined line width.
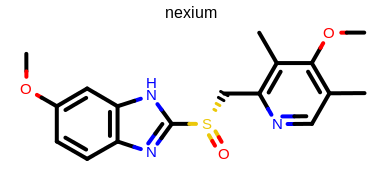
Example of using the program with user-defined line width
prompt> Mol2Img -in nexium.ism -out image.png -superdisp All
will generate the image shown in Figure: Example of using the program with superatom style.
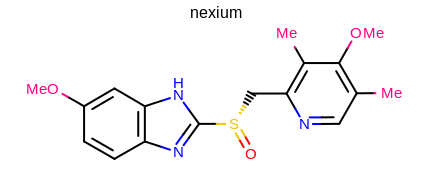
Example of using the program with superatom style
Note
The above program can generate 2D coordinates with user-defined 2D ring layouts when using the -ringdict parameter.
See also the 2D Coordinate Generation Examples section of the OEChem TK manual that shows examples about how to generate and utilize user-defined ring dictionaries.
See also
Molecule Depiction chapter