Version 2.0.0
OEChem 2.0.0
New features
OEMolDatabase class added for providing fast random read-only access to all the file formats OEChem supports. Please see the new Molecular Database Handling chapter of the documentation for a more thorough description.
Warning
The meaning of the .smi file extension has changed in
OEChem 2.0. The .smi file extension will now retain
stereochemistry information. This could cause problems for systems
that relied upon .smi to strip away stereo chemistry.
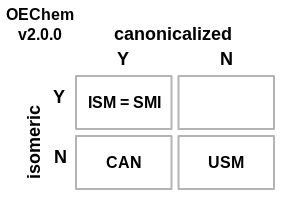
The following changes have been made to how OEChem defines the various flavors of SMILES (see images demonstrating these changes in Table: SMILES File Formats):
The
.smi,OEFormat.SMI, file format outputs Canonical isomeric SMILES.OEFormat.USMfile format added (with the.usmfile extension) that allows the generation of non-canonical, non-isomeric SMILES, i.e., the OEChem 1.x definition of the.smifile extension.OEGetFormatStringreturns more descriptive values for the various flavors of SMILES regarding canonicalization and stereo information.The default file format for molecule streams is now
OEFormat.SMIand is identical to theOEFormat.ISMformat.
SMILES File Formats 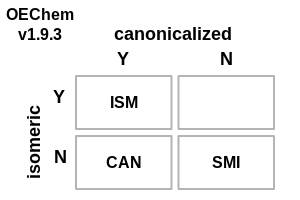
OEFormat.CSVfile format added to OEChem for round-tripping molecules and SD data to other software packages that support the comma-separate-value,.csv, file format. OEChem supports the.csvfile format that has largely become ubiquitous and is now standardized by RFC 4180. The following APIs were added for handling CSV files in OEChem:Added automatic 2D coordinate generation to the following file formats when invoking the
OEWriteMoleculehigh-level molecule writer:In case of the
OEFormat.MDLand theOEFormat.SDFfile formats, 2D coordinates are generated if the molecule has no coordinates i.e. existing 2D or 3D coordinates will be left intact.In case of the
OEFormat.CDXfile format, 2D coordinates are generated if the molecule has no 2D coordinates.
This default behavior can be turned off by using the
OEOFlavor.MDL.Add2D,OEOFlavor.SDF.Add2DandOEOFlavor.CDX.Add2Dflags, respectively.OEGetFormatExtensionwill now return the most common file extension as the first element in the comma separated list for theOEFormat.PDB,OEFormat.SDF, andOEFormat.RDFfile formats.OEGenerate2DCoordinatesadded to assign 2D coordinates to the given molecule.OEMatchBase.IsValidandoperator boolmethods added.OERoleSet abstract base class added as a mixin class to allow a class to contain a set of OERole objects for classification purposes. This is similar to how the OEBase class provides associative data container behavior as “generic data”. The following classes already derive from OERoleSet:
More classes across the toolkits may be added in the future as dictated by needs.
oemolstreambase.GetFileNamemethod added to all molecule streams to return the file name used to open the stream, if a file name was used.OEMatchBase.IsValidmethod added for determining whether the match contains any atoms or bonds.OEAssignZap7Radiifunction added to assign radii from the ZAP7 set.OEParseSmilesOptionsclass added for adding more complex SMILES parsing options for theOEParseSmilesfunction. This added the ability to makeOEParseSmilesquiet with regard to parsing failures through theOEParseSmilesOptions.SetQuietmethod.
Major bug fixes
The
OEFormat.MDLV3000 file format was unable to read multiple molecules from the same file if they were all in the V3000 format. Note, this did not affect theOEFormat.SDFfiles containing V3000 as those files contain ‘$ $ $ $’ to delimit separate molecule records.OEChem::OEReadHeaderwill no longer corrupt the stack by arbitrarily zeroing out bytes on the stack whenever no OEHeader record is found in theOEFormat.OEBfile.OEMatch.AddPairwill no longer crash ifOEMatch.Clearwas previously called.oemolthreadbase.PutMolwill now destroy the pointer passed to it if the underlying buffer returnsfalse, e.g., the buffer has already been closed.OEAddMols,OESuppressHydrogens, andOEPerceiveSymmetrywill no longer cause a stack overflow and crash when the molecule has a large number of atoms.
A subtle memory and performance bug was fixed for the OEMol, OEGraphMol, and OEQMol copy constructors. Every time a copy was made an extra layer of proxy object was created, causing memory consumption to increase, as well as making any subsequent use of that object slower.
Minor bug fixes
Removed unimplemented
OEChem::OEFormat::TDTfile format.Removed
OEChem::OEFormat::BINformat after being deprecated for 10 years.OEMolToSmilesno longer outputs the title of the molecule.If
OE3DToAtomStereoorOE3DToBondStereothrows a warning message during a call toOEMolToSmiles, the warning message will no longer erroneously say it is during a call toOEWriteMolecule.OESetCommentandOEGetCommenthave have been slightly optimized for speed. There is also a larger optimization for memory and file space in the.oebformat. Previously, setting the comment to an empty string would write superfluous data to the.oebfile.OECopySDDataandOECopyPDBDatawill no longer increase the memory consumption of the destination molecule whenever the source molecule does not contain any data. This was causingOEFormat.PDBfiles read into a OEMCMolBase to use more memory than necessary.OEFormat.MOL2parser will no longer create bonds with zero-order from “dummy bonds”. Zero-order bonds cause problems for many OEChem algorithms likeOEKekulize.OEFormat.MOL2parser will now properly ignore lines between molecule records that start with the pound sign, “#”. Previously, these lines would cause OEChem to spew a lot of warnings and cause the parser to fail.
Documentation fixes
The chapter about InChI failures has been removed since most of the InChI failures were fixed in the last release, OEChem 1.9.3 in 2013.Oct.
Release notes section re-organized to make the current release more prominent.
OESystem 2.0.0
New features
Added support for handling the comma-separated-value, CSV, format specified by RFC 4180 with the following two free functions:
OEStringCSVJoinfor creating a CSV recordOEStringCSVTokenizefor parsing a CSV record
The following low-level functions were added support CSV handling but can be ignored by most users:
OEStringCSVQuoteaddedOEStringCollapseQuotesaddedOEStringStripQuotesadded
OEHalfFloat class added for storing floating point data as a 16-bit representation as specified by the IEEE 754-2008 standard to save on memory consumption and bandwidth.
The performance of reading plain-old-data attached to OEBase objects from
.oebfiles has been improved by about %10.OEErrorLevelToStringfree function added.OEBFPosTEndianadded for allowing 64-bit integers to be round-tripped to binary formats regardless of machine endian-ness.
Major bug fixes
OEThrowmutex handling has been migrated from OEErrorHandler down a level intoOESystem::OEErrorHandlerStreamImpl. This fixes the following issues:The mutex can now be properly released during a process exit like
OEErrorHandler.Fatal.A deadlock will no longer occur if the implementation of
OEErrorHandlerImplBase.Msgneeds to throw a message itself.Allows alternative faster and more scalable implementations of OEErrorHandlerImplBase to be created and used with
OEThrow.
Minor bug fixes
OEBitVectorNumWordsandOEBitVectorNumBytesnow take and returnsize_t.OESystem::OEBinaryTagMaxLengthconstant added and set to 1024, the maximum length of an explicit string tag in the.oebfile format.OEStringTokenizeQuotedwill no longer treat a quote as the end quote of a field if it is escaped by another quote. Table: OEStringTokenizeQuoted Change demonstrates the change to support proper CSV parsing.OEStringTokenizeQuoted Change OEChem 1.x
“foo””,bar”,blah
"foo""``|``bar"``|``blahOEChem 2.0+
“foo””,bar”,blah
"foo"",bar"``|``blahThe size of
OEBitVectorobject has been increased from 12 bytes to 16 bytes on 64-bit machines. The size is still 8 bytes on a 32-bit machine.
The OEErrorHandlerImplBase copy constructor is now usable in Python, Java, and C#.
Documentation fixes
IsTrue and IsFalse are deprecated and will be removed in
OEChem 3.0, please convert to using OEIsTrue and OEIsFalse.OEUnaryTrue and OEUnaryFalse documented as acceptable synonyms of OEIsTrue and OEIsFalse.
OEPlatform 2.0.0
New features
OEAddLicenseFromHttpadded to license the current process with an OpenEye license file retrieved from a valid URL. The only supported URL schemes are HTTP and HTTPS. A reference implementation of a license server can be found at: (https://github.com/oess/HttpLicenseServer).
OEAddLicenseDatafunction added to parse a string as if it is an OpenEye license file and then license the current process with it.
Major bug fixes
OEPlatform::oeistream::size was incorrectly being truncated to 32-bits on 32-bit machines. This resulted in incorrect file sizes being reported for files over 4 gigabytes in size on 32-bit machines.
OEPlatform::oeifstream::tell would return incorrect values if called after OEPlatform::oeifstream::size and after some bytes were already read from the stream.
Minor bug fixes
OEPlatform::OEMallocaPtr::GetPtradded to allow the object to be explicitly converted to a pointer type.OEMutex and OETryMutex destructors will now destroy themselves on
pthreadbased systems. Destroying mutexes is optional according to thepthreadstandard, but helpful in debugging possible deadlocks.OEMutex and OETryMutex no longer use
gthreads, instead usingpthreads, allowing for integration with non-GCC systems like libc++ on OSX.
OEGrid 1.4.5
Major bug fixes
OEGridFileType.Asciifile writer will no longer sometimes corrupt the stack and crash.When reading a CCP4 file, the standard deviation stored in the CCP4 header is now used to normalize the file.
OENormalizeGrid now properly normalizes by sigma (not variance)
Writing CCP4 maps now uses the original map statistics when possible.
Rotated skew grids attached as generic data are now round-trippable when saved to OEB
Fixed a crash when interpolating grids where the rotation matrix inverts the target grids bounding box
MTZ files with more than 18 columns are now read properly
Fixed a memory leak in
OESequenceAlignment.The constructor for the predicate OEHasResidueNumber now takes an int rather than an unsigned int because residue numbers can be negative.