OE2DMolDisplayOptions
class OE2DMolDisplayOptions
This class represents the OE2DMolDisplayOptions class that encapsulates properties that determine how a molecule is depicted.
The OE2DMolDisplayOptions class stores the following properties:
Property |
Get method |
Set method |
Corresponding namespace / class / type |
|---|---|---|---|
aromatic style |
|||
atomic number colors |
|||
atom color style |
|||
atom label font |
|||
atom label font scale |
positive floating point number in the range of |
||
atom property font |
|||
atom property label font scale |
positive floating point number in the range of |
||
atom property initializer |
|||
atom stereo style |
|||
atom SVG markup |
|||
atom visibility initializer |
|||
background color |
|||
bond color style |
|||
bond line gap scale |
positive floating point number in the range of |
||
bond line atom label gap scale |
positive floating point number in the range of |
||
bond property font |
|||
bond property label font scale |
positive floating point number in the range of |
||
bond property initializer |
|||
bond stereo style |
|||
bond pen |
|||
bond width scaling |
boolean |
||
bond SVG markup |
|||
explicit atom label angle |
positive floating point number in the range of |
||
height |
positive floating point number |
||
hydrogen style |
|||
margin(s) |
positive floating point number in the range of |
||
scale |
positive floating point number |
||
super atom label font |
|||
super atom display style |
|||
protective group label font |
|||
protective group display style |
|||
title font |
|||
title font scale |
positive floating point number in the range of |
||
title height |
|||
title location |
|||
width |
positive floating point number |
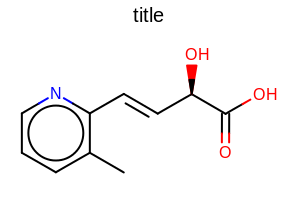
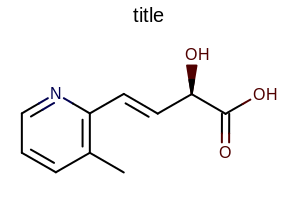
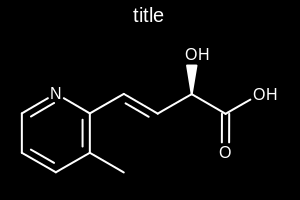
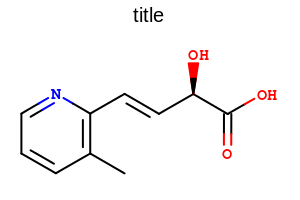
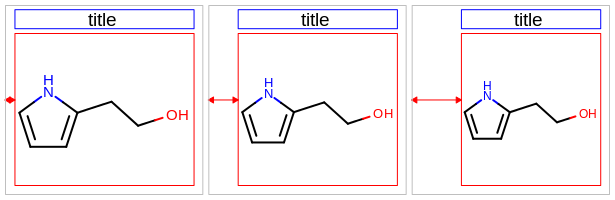
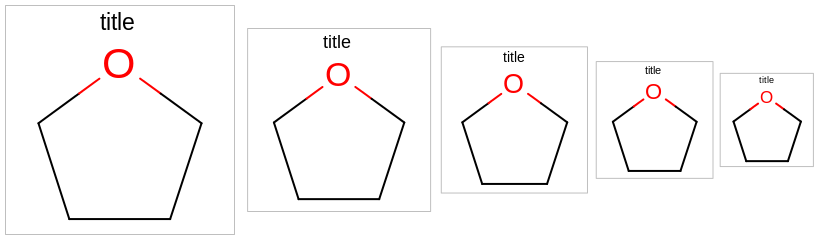
The images below illustrate the three possible molecule display layouts depending on the position of the title. These layouts can be controlled by the following parameters:
The width and the height can be set either by the constructor of the
OE2DMolDisplayOptionsclass, by calling theOE2DMolDisplayOptions.SetDimensionsmethod or by theSetWidthandSetHeightmethods, respectively.The location of the title area (blue box) can be set by the
SetTitleLocationmethod. The height of the title area (blue arrows) can be modified by invoking theSetTitleHeightmethod.The padding around the molecule (red arrows) can be changed by calling either the
SetMarginor theSetMarginsmethods.
title hidden |
title at the top |
title at the bottom |
|---|---|---|
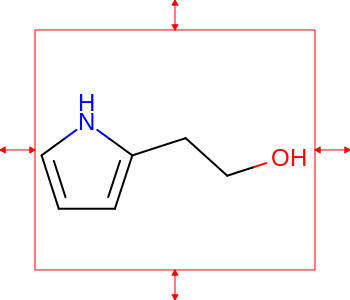
|
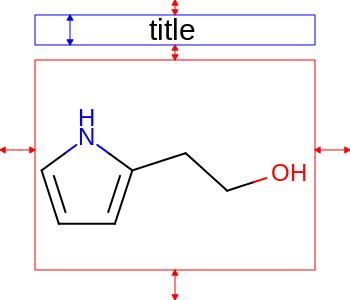
|
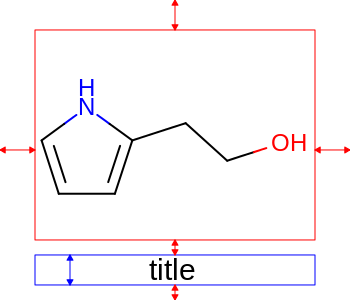
|
See also
OE2DMolDisplay class
Hint
Even though OEDepict TK provides access to manipulate the properties of the atom and bond displays after the OE2DMolDisplay object is constructed, it is highly recommended to determine the style of the depiction by using the OE2DMolDisplayOptions class. Only by knowing all properties (such as labels, font styles and sizes etc.) in advance can ensure the best depiction layout i.e. that the molecule diagram is rendered without any label clippings and the labels are displayed with the minimum number of overlaps.
Constructors
OE2DMolDisplayOptions()
Default constructor that initializes an OE2DMolDisplayOptions object with the following properties:
See example in Molecule depiction with default options)
Property |
Default value |
|---|---|
aromatic style |
|
atomic number colors |
initialized based on |
atom color style |
|
atom label font |
|
atom label font scale |
1.0 |
atom property font |
OEFont(OEColor(75, 75, 75)) |
atom property label font scale |
1.0 |
atom property initializer |
|
atom stereo style |
|
atom SVG markup |
|
atom visibility initializer |
|
background color |
|
bond color style |
|
bond line gap scale |
1.0 |
bond line atom label gap scale |
1.0 |
bond property font |
OEFont(OEColor(75, 75, 75)) |
bond property label font scale |
1.0 |
bond property initializer |
|
bond stereo style |
|
bond pen |
|
bond width scaling |
false |
bond SVG markup |
|
explicit atom label angle |
170.0 (degree) |
height |
0.0 (initialized based on scale) |
hydrogen style |
|
margins |
left and right margin 5.0% of the width; top and bottom margin 5.0% of the height of the molecule display |
scale |
|
super atom label font |
OEFont(OEPink) |
super atom display style |
|
protective group label font |
OEFont(OEPink) |
protective group display style |
|
title font |
|
title font scale |
1.0 |
title location |
|
title height |
10% of the height of the molecule display |
width |
0.0 (initialized based on scale) |
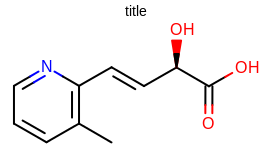
Molecule depiction with default options
OE2DMolDisplayOptions(double scale)
Creates an OE2DMolDisplayOptions object with the specified scaling.
- scale
A non-negative number that controls the magnification of the depicted molecule.
OE2DMolDisplayOptions(double width, double height, double scale)
Creates an OE2DMolDisplayOptions object with the specified dimension and scaling.
OE2DMolDisplayOptions(const OE2DMolDisplayOptions &rhs)
Copy constructor.
operator=
OE2DMolDisplayOptions &operator=(const OE2DMolDisplayOptions &rhs)
Assignment operator.
GetAromaticStyle
unsigned int GetAromaticStyle() const
Returns the style that controls how aromatic rings are displayed.
The return value is taken from the OEAromaticStyle
namespace.
See also
OEAromaticStylenamespace
GetAtomSVGMarkupFunctor
const OEAtomSVGMarkupBase &GetAtomSVGMarkupFunctor() const
Returns the functor that defines how atoms are marked in
svg image.
By default, residues are not marked (OEAtomSVGNoMarkup).
See also
GetAtomColor
const OESystem::OEColor &GetAtomColor(unsigned int atomic) const
Returns the color associated with a specific atomic number.
See also
OEColor class
Appendix: Element coloring (CPK) section
GetAtomColorStyle
unsigned int GetAtomColorStyle() const
Returns the style that is used to define the color of the
background and the default color of atom labels.
The return value is taken from the OEAtomColorStyle
namespace.
See also
OEAtomColorStylenamespace
GetAtomLabelFont
const OEFont &GetAtomLabelFont() const
Returns the font that is used to initialize the depiction style
of the atom labels
(OE2DAtomDisplay.GetLabelFont).
See also
OEFont class
GetAtomLabelFontScale
double GetAtomLabelFontScale() const
Returns the multiplier that can be used to increase or decrease the size of the atom label fonts.
See also
GetAtomPropLabelFont
const OEFont &GetAtomPropLabelFont() const
Returns the font that is used to initialize the depiction style
of the atom property labels
(OE2DAtomDisplay.GetPropertyFont).
See also
OEFont class
GetAtomPropLabelFontScale
double GetAtomPropLabelFontScale() const
Returns the multiplier that can be used to increase or decrease the size of the atom property label fonts.
See also
GetAtomPropertyFunctor
const OEDisplayAtomPropBase &GetAtomPropertyFunctor() const
Returns the functor that is used to initialize the atom property
labels (OE2DAtomDisplay.GetProperty).
See also
OEDisplayAtomPropBase class
OEDisplayAtomIdx class
OEDisplayAtomMapIdx class
OEDisplayNoAtomProp class
GetAtomVisibilityFunctor
const OESystem::OEUnaryPredicate<OEChem::OEAtomBase> &GetAtomVisibilityFunctor() const
Returns the functor that is used to identify visible atoms. The default
functor is the OEIsTrueAtom class which
results in all atoms being displayed.
See also
GetAtomStereoStyle
unsigned int GetAtomStereoStyle() const
Returns the style that controls what atom stereo information is displayed.
The return value is taken from the OEAtomStereoStyle
namespace.
See also
OEAtomStereoStylenamespace
GetBackgroundColor
const OESystem::OEColor &GetBackgroundColor() const
Returns the color that is used to clear the background of an image
(by calling OEImageBase.Clear method)
before rendering the molecule.
See also
OEColor class
OERenderMoleculefunction
GetBondColorStyle
unsigned int GetBondColorStyle() const
Returns the style that controls how bonds are colored.
The return value is taken from the OEBondColorStyle
namespace.
See also
OEBondColorStylenamespace
GetBondLineGapScale
double GetBondLineGapScale() const
Returns the multiplier that can be used to increase or decrease the gap between the lines of double and triple bonds.
See also
GetBondLineAtomLabelGapScale
double GetBondLineAtomLabelGapScale() constant
Returns the multiplier that can be used to increase or decrease the gap between the line(s) of bonds and the adjacent atom labels.
See also
GetBondPropLabelFont
const OEFont &GetBondPropLabelFont() const
Returns the font that is used to initialize the depiction style
of the bond property labels
(OE2DBondDisplay.GetPropertyFont).
See also
OEFont class
GetBondPropLabelFontScale
double GetBondPropLabelFontScale() const
Returns the multiplier that can be used to increase or decrease the size of the bond property label fonts.
See also
GetBondPropertyFunctor
const OEDisplayBondPropBase &GetBondPropertyFunctor() const
Returns the functor that is used to initialize the bond property
labels (OE2DBondDisplay.GetProperty).
See also
OEDisplayBondPropBase class
OEDisplayBondIdx class
OEDisplayNoBondProp class
GetBondStereoStyle
unsigned int GetBondStereoStyle() const
Returns the style that controls what bond stereo information is displayed.
The return value is taken from the OEBondStereoStyle
namespace.
See also
OEBondStereoStylenamespace
GetBondSVGMarkupFunctor
const OEBondSVGMarkupBase &GetBondSVGMarkupFunctor() const
Returns the functor that defines how bonds are marked in
svg image.
By default, residues are not marked (OEBondSVGNoMarkup).
See also
GetBondWidthScaling
bool GetBondWidthScaling() const
Returns whether the line width of the bond are increase or decreased based
on the molecule scaling factor (OE2DMolDisplayOptions.GetScale)
See also
GetDefaultBondPen
const OEPen &GetDefaultBondPen() const
Returns the pen that is used to initialize the bond pens
(OE2DBondDisplay.GetBgnPen and
OE2DBondDisplay.GetBgnPen).
See also
OEPen class
GetExplicitAtomLabelAngle
double GetExplicitAtomLabelAngle() const
In the case of an almost linear X-C-X bond, an explicit “C” label will be drawn if the angle is more that the returned value of this function. 180.0 degree means that the “C” label will never be drawn.
See also
GetHeight
double GetHeight() const
Returns the vertical limit into which the molecule has to be fitted.
See also
GetHydrogenStyle
unsigned int GetHydrogenStyle() const
Returns the style that controls of how implicit and explicit
hydrogens are displayed.
The return value is taken from the OEHydrogenStyle
namespace.
See also
OEHydrogenStylenamespace
GetMargin
double GetMargin(unsigned int margin) const
Returns the ratio of a specific margin of the OE2DMolDisplayOptions object.
- margin
This value has to be from the
OEMarginnamespace.
See also
OEMarginnamespace
GetProtectiveGroupLabelFont
const OEFont &GetProtectiveGroupLabelFont() const
Returns the font that is used to display the protective group labels.
See also
GetProtectiveGroupStyle
unsigned int GetProtectiveGroupStyle() const
Returns the style that controls whether or not specific pre-defined
protective group are contracted and labeled with corresponding
abbreviations.
The return value is taken from the OEProtectiveGroupStyle
namespace.
See also
Warning
This is a deprecated API.
Please use OE2DMolDisplayOptions.HasProtectiveGroupStyle
to determine which protective groups are set.
GetScale
double GetScale() const
Returns the scaling factor used to magnify a molecule.
See also
GetSuperAtomLabelFont
const OEFont &GetSuperAtomLabelFont() const
Returns the font that is used to display the abbreviations of contracted functional groups.
See also
GetSuperAtomStyle
unsigned int GetSuperAtomStyle() const
Returns the style that controls whether or not specific pre-defined
functional groups are contracted and labeled with corresponding
abbreviations.
The return value is taken from the OESuperAtomStyle
namespace.
See also
GetTitleFont
Returns the font that is used to display the title of the molecule
(i.e. the string returned by the OEMolBase.GetTitle
method).
const OEFont &GetTitleFont() const
See also
OEFont class
GetTitleFontScale
Returns the multiplier that can be used to increase or decrease the size of the title font.
double GetTitleFontScale() const
See also
GetTitleLocation
unsigned int GetTitleLocation() const
Returns the position of the molecule title.
The return value is taken from the OETitleLocation
namespace.
See also
OETitleLocationnamespace
GetTitleHeight
double GetTitleHeight() const
Returns the height of the title area.
See also
GetWidth
double GetWidth() const
Returns the horizontal limit into which the molecule has to be fitted.
See also
HasProtectiveGroupStyle
bool HasProtectiveGroupStyle(unsigned int style)
Returns true if the specific pre-defined protective group style is contracted and labeled with corresponding abbreviations.
- style
This value has to be from the
OEProtectiveGroupStylenamespace.
SetAromaticStyle
void SetAromaticStyle(unsigned int style)
Sets the style that controls how aromatic rings are displayed.
- style
This value has to be from the
OEAromaticStylenamespace.
Example: (Figure: Example of using the SetAromaticStyle method)
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAromaticStyle(OEAromaticStyle.Circle);
Example of using the SetAromaticStyle method
See also
OEAromaticStylenamespace
SetAtomColor
void SetAtomColor(unsigned int atomic, const OESystem::OEColor &color)
Sets the color associated with a specific atomic number.
- atomic
This value has to be from the
OEElemNonamespace of OEChem TK.- color
The color being associated with the given atomic number.
Example: (Figure: Example of using the SetAtomColor method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomColor(OEElemNo.O, new OEColor(80, 0, 0)); // very dark red
opts.SetAtomColor(OEElemNo.N, new OEColor(0, 0, 80)); // very dark blue
Example of using the SetAtomColor method
See also
OEColor class
Appendix: Element coloring (CPK) section
Note
The color of the atom also depends on the atom color style (
OE2DMolDisplayOptions.GetAtomColorStyle).Setting atom color style (
OE2DMolDisplayOptions.SetAtomColorStyle) to WhiteMonochrome sets white background and black color for all atoms. Likewise, setting atom color style to BlackMonochrome sets black background and white color for all atoms. But, user can choose to have another color for all atoms (in monochrome atom color style) by setting another atom color (OE2DMolDisplayOptions.SetAtomColor) for Carbon atoms. The example inListing 4demonstrates setting non-default atom colors in monochrome mode. Setting atom color for other than Carbon atoms sets color of only specified atom types, not for all atoms.
SetAtomColorStyle
void SetAtomColorStyle(unsigned int style)
Sets the style that is used to define the color of the background and the default color of atom labels.
- style
This value has to be from the
OEAtomColorStylenamespace.
Example: (Figure: Example of using the SetAtomColorStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomColorStyle(OEAtomColorStyle.BlackMonochrome);
Example of using the SetAtomColorStyle method
See also
OEAtomColorStylenamespace
SetAtomLabelFont
void SetAtomLabelFont(const OEFont &font)
Sets the font that is used to initialize the depiction style
of the atom labels
(OE2DAtomDisplay.GetLabelFont).
Example: (Figure: Example of using the SetAtomLabelFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetStyle(OEFontStyle.Bold);
opts.SetAtomLabelFont(font);
Example of using the SetAtomLabelFont method
Note
The color of the atom label fonts also depends on the atom color style (
OE2DMolDisplayOptions.GetAtomColorStyle).The size of fonts of the atom labels also depends on:
the scaling factor used to fit a molecule to a given dimension (
OE2DMolDisplayOptions.GetScale)the multiplier set by the
OE2DMolDisplayOptions.SetAtomLabelFontScalemethod
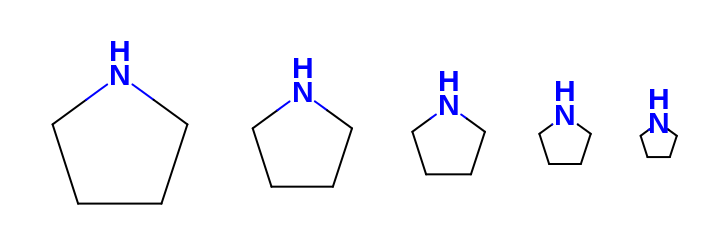
See example in Figure: Example of scaling the font of the atom label along with the molecule
Example of scaling the font of the atom label along with the molecule
See also
OEFont class
SetAtomLabelFontScale
void SetAtomLabelFontScale(double scale)
Sets the multiplier that can be used increase or decrease the size of the fonts of the atom labels.
- scale
This value has to be either
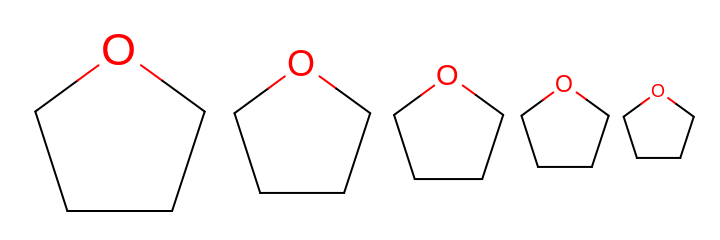
0.0or in a range of[0.5, 3.0]. See examples in Figure: Examples of scaling the font of the atom label relative to the moleculeIn case of
0.0, the atom labels are not scaled with the molecule but rather fixed font size are used regardless of the size of the molecule. See example in Figure: Example of atom labels with fixed font size
Examples of scaling the font of the atom label relative to the molecule
Example of atom labels with fixed font size
Example: (Figure: Example of using the SetAtomLabelFontScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomLabelFontScale(1.5);
Example of using the SetAtomLabelFontScale method
See also
SetAtomPropLabelFont
void SetAtomPropLabelFont(const OEFont &font)
Sets the font that is used to initialize the depiction style
of the atom property labels
(OE2DAtomDisplay.GetPropertyFont).
Example: (Figure: Example of using the SetAtomPropLabelFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetStyle(OEFontStyle.Bold);
font.SetColor(OEChem.OEDarkGreen);
opts.SetAtomPropLabelFont(font);
opts.SetAtomPropertyFunctor(new OEDisplayAtomIdx());
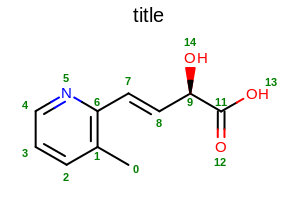
Example of using the SetAtomPropLabelFont method
Note
The size of the atom property label fonts also depends on the scaling factor used to fit a molecule to a given dimension (
OE2DMolDisplayOptions.GetScale) and the multiplier set by theOE2DMolDisplayOptions.SetAtomPropLabelFontScalemethod. See example in Figure: Example of scaling the font of the atom property labels along with the molecule
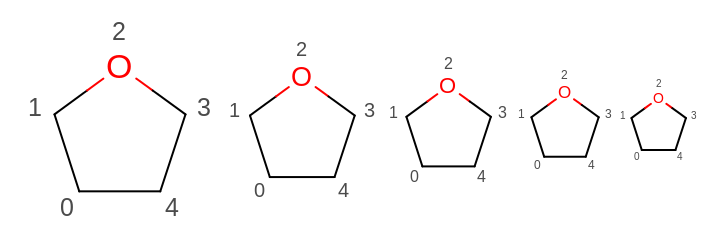
Example of scaling the font of the atom property labels along with the molecule
See also
OEFont class
SetAtomPropLabelFontScale
void SetAtomPropLabelFontScale(double scale)
Sets the multiplier that can be used increase or decrease the size of the fonts of the atom property labels.
- scale
This value has to be in the range of
[0.5, 2.0]. See examples in Figure: Examples of scaling the font of the atom property labels relative to the molecule
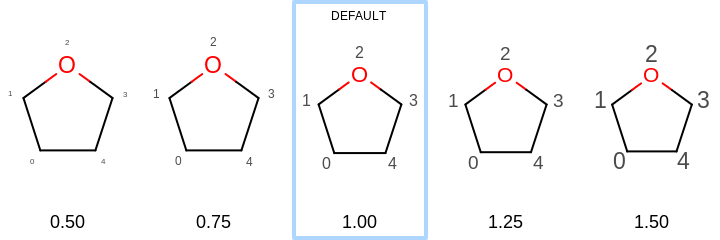
Examples of scaling the font of the atom property labels relative to the molecule
Example: (Figure: Example of using the SetAtomPropLabelFontScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomPropLabelFontScale(1.5);
opts.SetAtomPropertyFunctor(new OEDisplayAtomIdx());
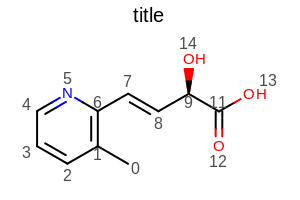
Example of using the SetAtomPropLabelFontScale method
See also
SetAtomPropertyFunctor
void SetAtomPropertyFunctor(const OEDisplayAtomPropBase &func)
Sets the functor that is used to initialize the atom property
labels (OE2DAtomDisplay.GetProperty)
Example: (Figure: Example of using the SetAtomPropertyFunctor method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomPropertyFunctor(new OEDisplayAtomIdx());
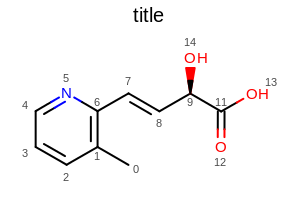
Example of using the SetAtomPropertyFunctor method
Hint
It is recommended to use the OE2DMolDisplayOptions.SetAtomPropertyFunctor
method to define the atom property labels when the OE2DMolDisplay
object is constructed.
All labels displayed on the molecule diagram have to be known in advance in order to be
able to minimize the number of label clashes and clippings when calculating
the positions of the bond property labels.
See example of user-defined atom properties in Displaying Atom Properties section.
See also
OEDisplayAtomPropBase class
OEDisplayAtomIdx class
OEDisplayAtomMapIdx class
OEDisplayNoAtomProp class
SetAtomStereoStyle
void SetAtomStereoStyle(unsigned int style)
Sets the style that controls what atom stereo information is displayed.
- style
This value has to be from the
OEAtomStereoStylenamespace.
Example: (Figure: Example of using the SetAtomStereoStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomStereoStyle(OEAtomStereoStyle.Display.All|OEAtomStereoStyle.HashWedgeStyle.Standard);
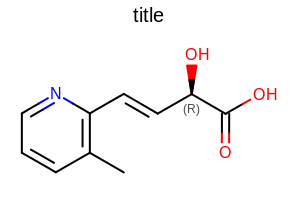
Example of using the SetAtomStereoStyle method
See also
OEAtomStereoStylenamespace
SetAtomSVGMarkupFunctor
void SetAtomSVGMarkupFunctor(const OEAtomSVGMarkupBase &func)
Sets the functor that defines how atoms are marked in svg image.
Drawing elements representing atoms in svg image are grouped together
in the following format in which the <group id> and <class name>
strings are defined by the given functor:
<g id='<group id>' class='<class name>'>
..
list of drawing elements
..
</g>
Note
This setting has only effect when generating .svg
images.
See also
OEAtomSVGMarkupBase abstract base class
OEAtomSVGNoMarkup class
OEAtomSVGAtomIdxMarkup class
OEAtomSVGResidueMarkup class
SetAtomVisibilityFunctor
void SetAtomVisibilityFunctor(const OESystem::OEUnaryPredicate<OEChem::OEAtomBase> &func)
Sets the functor that is used to identify visible atoms.
Example: (Figure: Example of using the SetAtomVisibilityFunctor method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetAtomVisibilityFunctor(new OEAtomIsInRing());
Example of using the SetAtomVisibilityFunctor method to only display rings
Hint
It is recommended to use the OE2DMolDisplayOptions.SetAtomVisibilityFunctor
method to identify the visible atoms when the OE2DMolDisplay
object is constructed. All objects displayed in the molecule diagram have to be known in advance in order to be
able to correctly size and scale the molecule.
See additional examples of atom visibility control in the Displaying Atom Properties section.
See also
SetBackgroundColor
void SetBackgroundColor(const OESystem::OEColor &color)
Sets the color that is used to clear the background of an image
(by calling OEImageBase.Clear method)
before rendering the molecule.
Example: (Figure: Example of using the SetBackgroundColor method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBackgroundColor(OEChem.OEYellowTint);
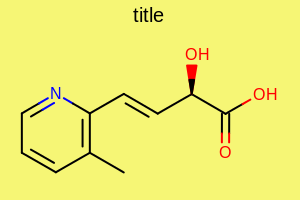
Example of using the SetBackgroundColor method
See also
OEColor class
SetBondColorStyle
void SetBondColorStyle(unsigned int style)
Sets the style that controls how bonds are colored.
- style
This value has to be from the
OEBondColorStylenamespace.
Example: (Figure: Example of using the SetBondColorStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondColorStyle(OEBondColorStyle.Monochrome);
Example of using the SetBondColorStyle method
See also
OEBondColorStylenamespace
SetBondLineGapScale
void SetBondLineGapScale(double scale)
Sets the multiplier that can be used increase or decrease the gap between the lines of double and triple bonds.
- scale
This value has to be in the range of
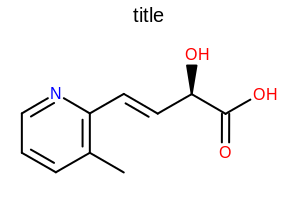
[0.5, 2.0]. See examples in Figure: Examples of altering the gap between the lines of double and triple bonds
Examples of altering the gap between the lines of double and triple bonds
Example: (Figure: Example of using the SetBondLineGapScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondLineGapScale(0.5);
Example of using the SetBondLineGapScale method
See also
SetBondLineAtomLabelGapScale
void SetBondLineAtomLabelGapScale(double scale)
Sets the multiplier that can be used to increase or decrease the gap between the line(s) of bonds and the adjacent atom labels.
- scale
This value has to be in the range of
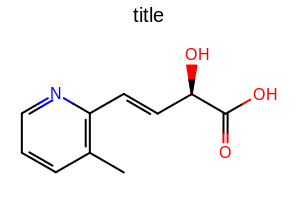
[0.5, 2.0]. See examples in Figure: Examples of altering the gap between the line(s) of bonds and adjacent atom labels
Examples of altering the gap between the line(s) of bonds and adjacent atom labels
Example: (Figure: Example of using the SetBondLineAtomLabelGapScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondLineAtomLabelGapScale(2.0);
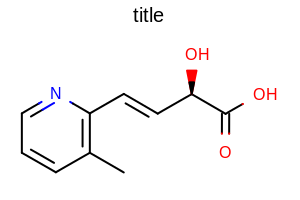
Example of using the SetBondLineAtomLabelGapScale method
See also
SetBondPropLabelFont
void SetBondPropLabelFont(const OEFont &color)
Sets the font that is used to initialize the depiction style
of the bond property labels
(OE2DBondDisplay.GetPropertyFont).
Example: (Figure: Example of using the SetBondPropLabelFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetStyle(OEFontStyle.Bold);
font.SetColor(OEChem.OEDarkGreen);
opts.SetBondPropLabelFont(font);
opts.SetBondPropertyFunctor(new OEDisplayBondIdx());
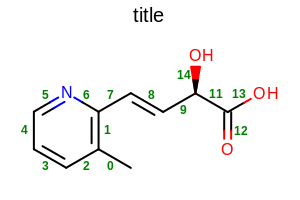
Example of using the SetBondPropLabelFont method
Note
The size of the bond property label fonts also depends on the scaling factor used to fit a molecule to a given dimension (
OE2DMolDisplayOptions.GetScale) and the multiplier set by theOE2DMolDisplayOptions.SetBondPropLabelFontScalemethod. See example in Figure: Example of scaling the font of the bond property labels along with the molecule
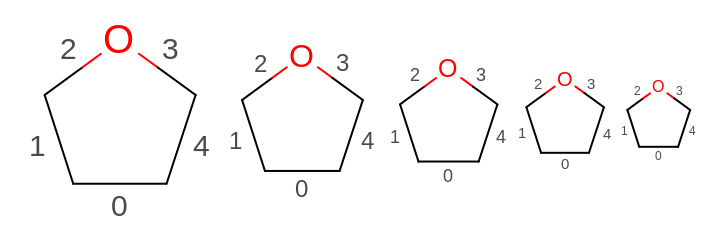
Example of scaling the font of the bond property labels along with the molecule
See also
OEFont class
SetBondPropLabelFontScale
void SetBondPropLabelFontScale(double scale)
Sets the multiplier that can be used increase or decrease the size of the fonts of the bond property labels.
- scale
This value has to be in the range of
[0.5, 2.0]. See examples in Figure:*Examples of scaling the font of the bond property labels relative to the molecule SetBondPropLabelFontScale method
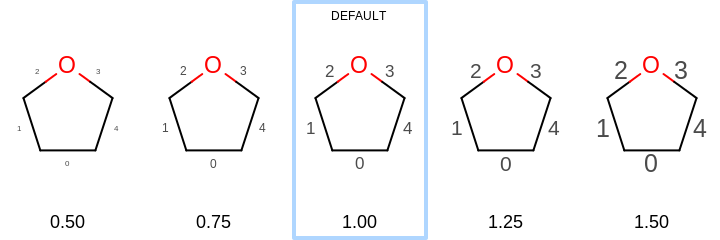
Examples of scaling the font of the bond property labels relative to the molecule
Example: (Figure: Example of using the SetBondPropLabelFontScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondPropLabelFontScale(1.5);
opts.SetBondPropertyFunctor(new OEDisplayBondIdx());
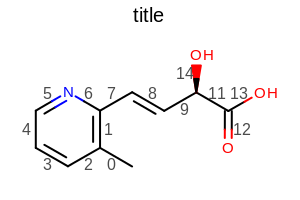
Example of using the SetBondPropLabelFontScale method
See also
SetBondPropertyFunctor
void SetBondPropertyFunctor(const OEDisplayBondPropBase &func)
Sets the functor that is used to initialize the bond property
labels (OE2DBondDisplay.GetProperty)
Example: (Figure: Example of using the SetBondPropertyFunctor method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondPropertyFunctor(new OEDisplayBondIdx());
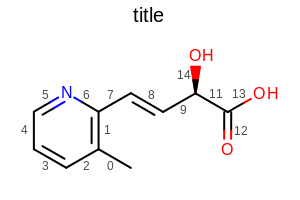
Example of using the SetBondPropertyFunctor method
Hint
It is recommended to use the OE2DMolDisplayOptions.SetBondPropertyFunctor
method to define the bond property labels when the OE2DMolDisplay
object is constructed.
All labels displayed on the molecule diagram have to be known in advance in order to be
able to minimize the number of label clashes and clippings when calculating
the positions of the bond property labels.
See example of user-defined bond properties in Displaying Bond Properties section.
See also
OEDisplayBondPropBase class
OEDisplayBondIdx class
OEDisplayNoBondProp class
SetBondStereoStyle
void SetBondStereoStyle(unsigned int style)
Sets the style that controls what bond stereo information is displayed.
- style
This value has to be from the
OEBondStereoStylenamespace.
Example: (Figure: Example of using the SetBondStereoStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondStereoStyle(OEBondStereoStyle.Display.All);
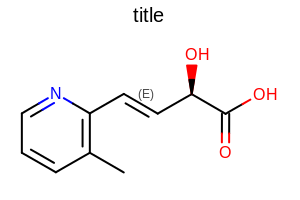
Example of using the SetBondStereoStyle method
See also
OEBondStereoStylenamespace
SetBondSVGMarkupFunctor
void SetBondSVGMarkupFunctor(const OEBondSVGMarkupBase &func)
Sets the functor that defines how bonds are marked in svg image.
Drawing elements representing bonds in svg image are grouped together
in the following format in which the <group id> and <class name>
strings are defined by the given functor:
<g id='<group id>' class='<class name>'>
..
list of drawing elements
..
</g>
Note
This setting has only effect when generating .svg
images.
See also
OEBondSVGMarkupBase abstract base class
OEBondSVGNoMarkup class
OEBondSVGBondIdxMarkup class
SetBondWidthScaling
void SetBondWidthScaling(bool scale)
Sets whether the line width of the bond are increase or decreased based
on the molecule scaling factor (OE2DMolDisplayOptions.GetScale).
See examples in Figure: Examples of bond line width.
Note
The line width of the bonds also depends on the default bond pen that can be set by using the
OE2DMolDisplayOptions.SetDefaultBondPenmethod.
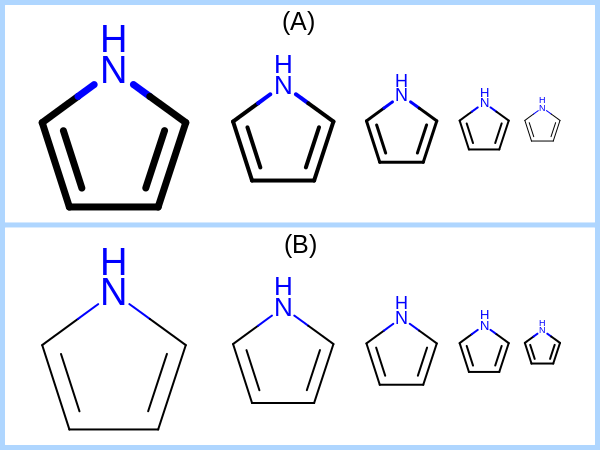
Examples of bond line width: (A) scaling the line width of the bonds relative to the molecule; (B) using constant line width that does not depend on the molecule scaling
Example: (Figure: Example of using the SetBondWithScaling method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetBondWidthScaling(true);
Example of using the SetBondWidthScaling method
See also
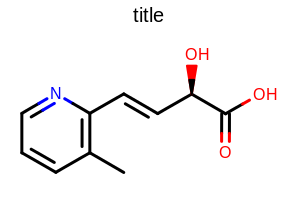
SetDefaultBondPen
void SetDefaultBondPen(const OEPen &pen)
Sets the pen that is used to initialize the bond pens
(OE2DBondDisplay.GetBgnPen and
OE2DBondDisplay.GetEndPen)
Example: (Figure: Example of using the SetDefaultBondPen method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEPen pen = new OEPen(OEChem.OEBlack, OEChem.OEBlack, OEFill.On, 5.0);
opts.SetDefaultBondPen(pen);
Example of using the SetDefaultBondPen method
Note
The style of the pens being used to draw bonds also depends on the bond display style (OE2DBondDisplay)
The color of the pens being used to draw bonds also depends on the given atom and bond color styles (
OE2DMolDisplayOptions.SetAtomColorStyleandOE2DMolDisplayOptions.SetBondColorStyle, respectively)
See also
OEPen class
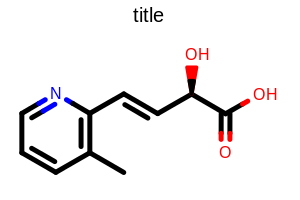
SetDimensions
void SetDimensions(double width, double height, double scale)
Sets parameters that control the size of the depicted molecule.
- width
A non-negative number that controls the horizontal limit.
- height
A non-negative number that controls the vertical limit.
- scale
A non-negative number that controls the magnification of the depicted molecule.
OEScale.AutoScalescaling means that the molecule is scaled to maximally fit the given dimensions.
Example: (Figure: Example of using the SetDimensions method)
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions();
double width = 180.0;
double height = 180.0;
double scale = 20.0;
opts.SetDimensions(width, height, scale);
Example of using the SetDimensions method
See also
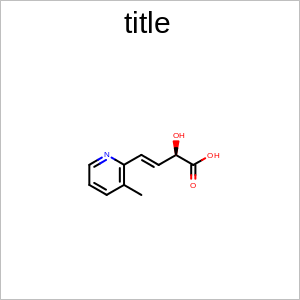
SetExplicitAtomLabelAngle
void SetExplicitAtomLabelAngle(const double angle)
In the case of an almost linear X-C-X bond, an explicit “C” label will be drawn if the angle is more that the given angle. 180.0 degree means that the “C” label will never be drawn.
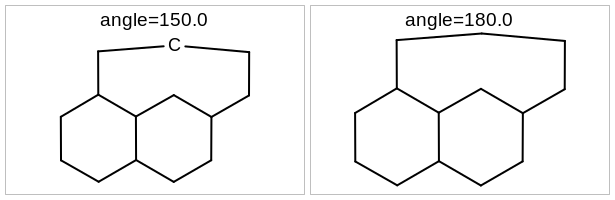
Example of using the SetExplicitAtomLabelAngle method
See also
SetHeight
void SetHeight(double height)
Sets the vertical limit into which the molecule has to be fitted.
- height
This number has to be non-negative number.
Example: (Figure: Example of using the SetHeight method)
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions();
opts.SetHeight(150.0);
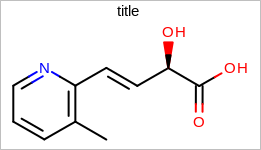
Example of using the SetHeight method
See also
SetHydrogenStyle
void SetHydrogenStyle(unsigned int style)
Sets the style that controls how implicit and explicit hydrogens are displayed.
- style
This value has to be from the
OEHydrogenStylenamespace.
Example: (Figure: Example of using the SetHydrogenStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetHydrogenStyle(OEHydrogenStyle.ImplicitTerminal);
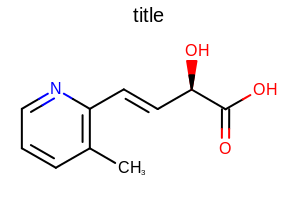
Example of using the SetHydrogenStyle method
See also
OEHydrogenStylenamespace
SetMargin
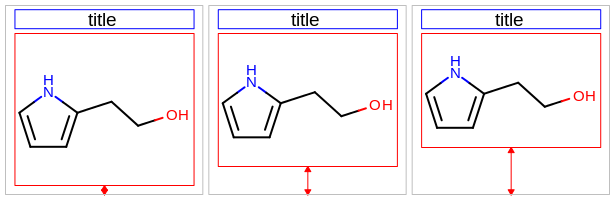
void SetMargin(unsigned int marginloc, double margin)
Sets the size of a specific margin of the OE2DMolDisplayOptions object.
- marginloc
This value has to be from the
OEMarginnamespace.- margin
This number is considered as a percentage of either the width or the height of the molecule display and has to be in the range of
[2.5%, 25.0%]. See example in Table: Example of setting specific margins to 5% (default), 15% and 25%. For example, when setting a margin to 15.0%:in case of the top or the bottom margins, it means that the margin will be 15% of the total height of the molecule display
in case of the left or the right margins, it means that the margin will be 15% of the total width of the molecule display
top margin |
bottom margin |
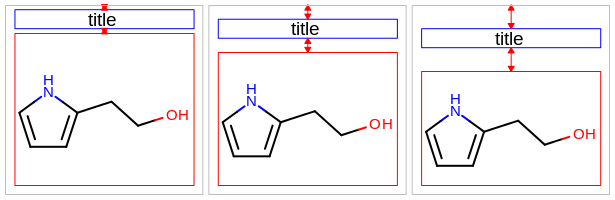
|
|
left margin |
right margin |
|
|
Example: (Figure: Example of using the SetMargin method)
double width = 240.0;
double height = 180.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetMargin(OEMargin.Bottom, 25.0);
Example of using the SetMargin method
See also
OEMarginnamespace
SetMargins
void SetMargins(double margin)
Sets the size of all margins of the OE2DMolDisplayOptions object.
- margin
This number is considered as a percentage of either the width or the height of the molecule display and has to be in the range of
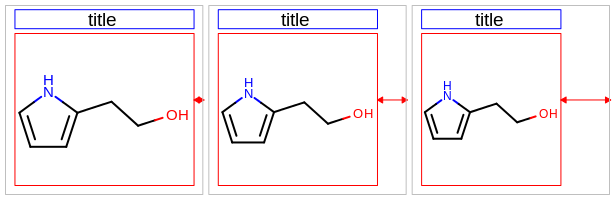
[2.5%, 25.0%]. For example, 15.0% means, that the left and right margin are 15% of the total width of the molecule display, and the top and bottom margins are 15% of the total height of the molecule display. See example in Figure: Example of setting all margins to 5% (default), 15% and 25%.
Example of setting all margins to 5% (default), 15% and 25%
Example: (Figure: Example of using the SetMargin method)
double width = 240.0;
double height = 180.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetMargins(25.0);
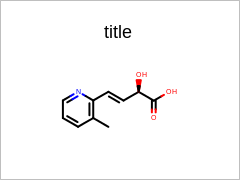
Example of using the SetMargins method
See also
OEMarginnamespace
SetProtectiveGroupLabelFont
void SetProtectiveGroupLabelFont(const OEFont &font)
Sets the font that is used to display the abbreviations of contracted functional groups.
Example: (Figure: Example of using the ProtectiveGroupLabelFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetColor(OEChem.OEDarkRed);
opts.SetProtectiveGroupLabelFont(font);
opts.SetProtectiveGroupStyle(OEProtectiveGroupStyle.All);
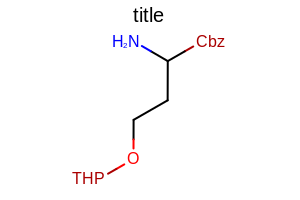
Example of using the SetProtectiveGroupLabelFont method
See also
Note
The size of the protective group fonts also depends on the scaling factor used to fit a molecule to a given dimension (
OE2DMolDisplayOptions.SetScale).
SetProtectiveGroupStyle
void SetProtectiveGroupStyle(unsigned int style)
Sets the style that controls whether or not specific pre-defined protective group are contracted and labeled with corresponding abbreviations.
- style
This value has to be from the
OEProtectiveGroupStylenamespace.
Example: (Figure: Example of using the SetProtectiveGroupStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetProtectiveGroupStyle(OEProtectiveGroupStyle.All);
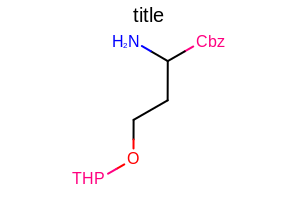
Example of using the SetProtectiveGroupStyle method
See also
SetScale
void SetScale(double scale)
Sets the scaling factor used to magnify a molecule.
- scale
This number has to be non-negative number.
Example: (Figure: Example of using the SetScale method)
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions();
opts.SetScale(20.0);
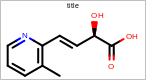
Example of using the SetScale method
See also
SetSuperAtomLabelFont
void SetSuperAtomLabelFont(const OEFont &font)
Sets the font that is used to display the abbreviations of contracted functional groups.
Example: (Figure: Example of using the SetSuperAtomLabelFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetColor(OEChem.OEDarkRed);
opts.SetSuperAtomLabelFont(font);
opts.SetSuperAtomStyle(OESuperAtomStyle.All);
Example of using the SetSuperAtomLabelFont method
See also
OEFont class
Note
The size of the super atom fonts also depends on the scaling factor used to fit a molecule to a given dimension (
OE2DMolDisplayOptions.SetScale).
SetSuperAtomStyle
void SetSuperAtomStyle(unsigned int style)
Sets the style that controls whether or not specific pre-defined functional groups are contracted and labeled with corresponding abbreviations.
- style
This value has to be from the
OESuperAtomStylenamespace.
Example: (Figure: Example of using the SetSuperAtomStyle method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetSuperAtomStyle(OESuperAtomStyle.All);
Example of using the SetSuperAtomStyle method
See also
OESuperAtomStylenamespace
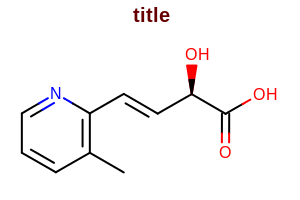
SetTitleFont
void SetTitleFont(const OEFont &font)
Sets the font that is used to display the title of the molecule
(i.e. the string returned by the OEMolBase.GetTitle
method).
Example: (Figure: Example of using the SetTitleFont method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OEFont font = new OEFont();
font.SetColor(OEChem.OEDarkBrown);
font.SetStyle(OEFontStyle.Bold);
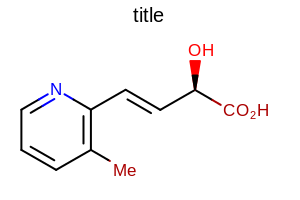
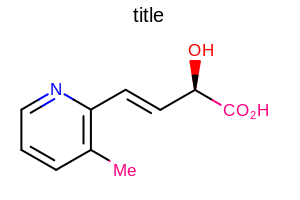
opts.SetTitleFont(font);
Example of using the SetTitleFont method
Note
The size of the title font also depends on:
the width and height of the molecule display
the height of the title area that can be set by
OE2DMolDisplayOptions.SetTitleHeightmethodthe multiplier that can be set by the
OE2DMolDisplayOptions.SetTitleFontScalemethod
See example in Figure: Example of scaling the title along with the molecule
Example of scaling the title along with molecule
See also
OEFont class
SetTitleFontScale
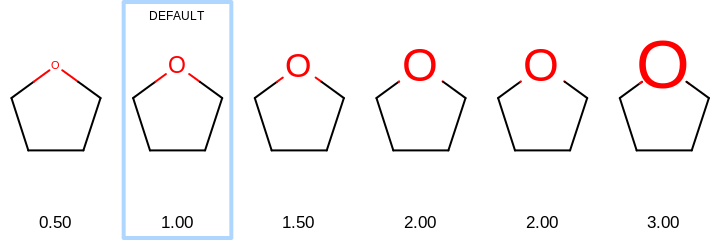
void SetTitleFontScale(double scale)
Sets the multiplier that can be used increase or decrease the size of the fonts of the atom labels.
- scale
This value has to be either
0.0or in a range of[0.5, 2.0]. See examples in Examples of scaling the font of title relative to the molecule In case of0.0, the title is not scaled with the molecule but rather fixed font size is used regardless of dimensions of the image. See example in Figure: Example of titles with fixed font size
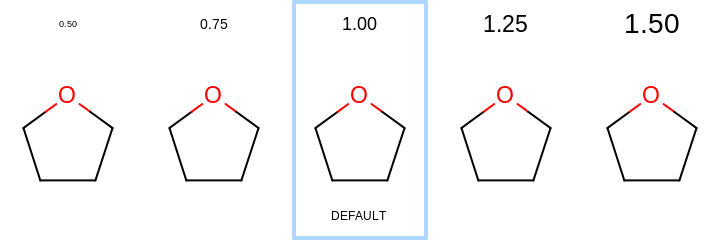
Examples of scaling the font of title relative to the molecule
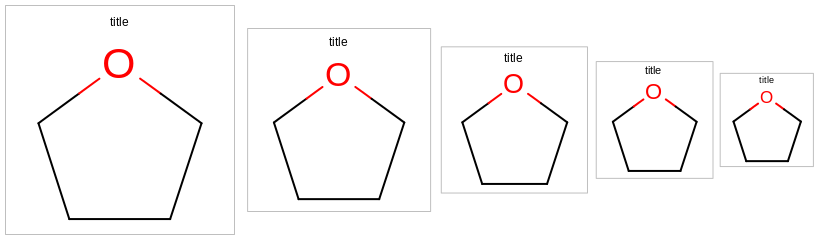
Example of titles with fixed font size
Example: (Figure: Example of using the SetTitleFontScale method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetTitleFontScale(0.5);
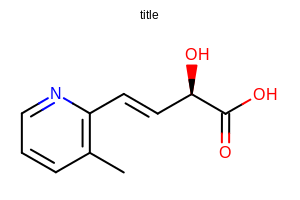
Example of using the SetTitleFontScale method
See also
SetTitleLocation
void SetTitleLocation(unsigned int loc)
Sets the position of the molecule title.
- loc
This value has to be from the
OETitleLocationnamespace.
Example: (Figure: Example of using the SetTitleLocation method)
double width = 300.0;
double height = 200.0;
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
opts.SetTitleLocation(OETitleLocation.Bottom);
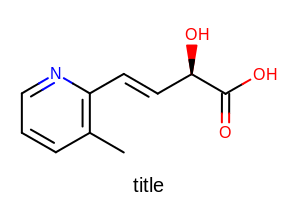
Example of using the SetTitleLocation method
See also
OETitleLocationnamespace
SetTitleHeight
void SetTitleHeight(double height)
Sets the height of the title area of the OE2DMolDisplayOptions object.
- height
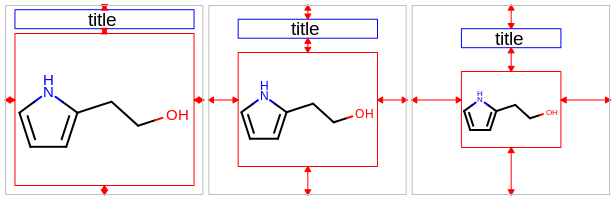
This number is considered as a percentage of the height of the molecule display and has to be in the range of
[5.0%, 25.0%]. See example in Figure: Example of setting height of the title area to 5%, 10% (default) and 25%.
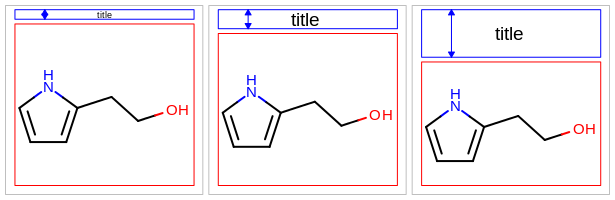
Example of setting height of the title area to 5%, 10% (default) and 25%
Note
A molecule display has a title area only if the title location is not
hidden. See also OETitleLocation namespace.
Example: (Figure: Example of using the SetTitleHeight method)
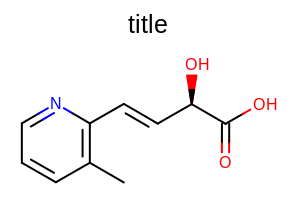
Example of using the SetTitleHeight method
See also
SetWidth
void SetWidth(double width)
Sets the horizontal limit into which the molecule has to be fitted.
- width
This number has to be non-negative number.
Example: (Figure: Example of using the SetWidth method)
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions();
opts.SetWidth(150.0);
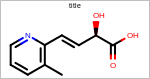
Example of using the SetWidth method
See also