Version 2.0.2
OEChem 2.0.2
New features
OEGenerate2DCoordinateshas an additional 550 complex ring systems that it can use while laying out coordinates. Any structures that contain one of these complex ring systems will see dramatically better 2D coordinate generation.OEChem::OEMCMolBaseTandOEChem::OEConfBaseThave been renamed to OEMCMolBase and OEConfBase respectively. This can only really have a technical effect on users of the direct C++ API. Python, Java, and C# users never saw a distinction between the template classes with theTsuffix. For higher level language users, navigating the documentation and OEChem’s type hierarchy should be greatly simplified.OEUncolorMolis a new function for performing tunable uncoloring strategies for an input molecule. Additional uncoloring strategies may be available in future versions.Significant performance improvement for
OEMolBase.GetAtoms. Algorithms that do molecular graph traversal should see a significant performance benefit. For example,OEShortestPathwas measured to be 25% faster.OEPreserveRotCompressadded as a convenience function to ensure rotor-offset-compression is preserved when reading and writing molecules to OEB files. This function should be called on any oemolistream object that should preserve the rotor-offset-compression data needed to reproduce a smaller OEB file.The performance of reading and writing rotor-offset-compression files has been improved, both when rotor-offset-compression data is preserved, and when it is not.
Multiple lines of a CSV file format can be read into successive conformers of a OEMCMolBase object. The SD data representing the data in the CSV file will be attached to each conformer.
OEAtomBondSet added the following new methods to support the addition and removal of atom and bond object from the set:
Adding of objects now verifies the membership of the object in the set prior to addition which will incur slightly more overhead than previous versions. This membership checking is in keeping with the spirit of uniqueness for a “set” implementation.
OESubsetMolhas new overloads to support use of subsetting activities using the OEAtomBondSet object to define the subset.OEMDLPerceiveBondStereowill causeOEReadMoleculeto produce fewer warning messages.OEMDLPerceiveBondStereowas creating ambiguous stereo bond marks, additional attempts are now made to apply unambiguous stereo bond marks. In some cases, this may result in >1 bond mark to a center to fulfill the requirements of the internal checking algorithm for validly marked centers. Pathological geometries remain where no valid marks can unambiguously define the stereocenter. For example, for some fused ring systems, the required (perhaps multiple) stereomarks would result in unpalatable marked stereocenters.Hint
For some cases,
OEMDLPerceiveBondStereowill not be able to apply marks that will satisfy theOEReadMoleculeperception, and a warning message will be thrown byOEReadMoleculeindicating that these should be redrawn or have coordinates regenerated, e.g., adding an explicit hydrogen at the bridgehead(s) to carry a stereomark.Note
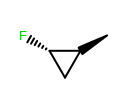
OEWriteMoleculeautomatically callsOEMDLPerceiveBondStereowhen writing to theOEFormat.MDLandOEFormat.SDFfile formats.OEMDLHasIncorrectBondStereohas been modified to check additional stereo environments for potential errors. For example, both example structures below are now identified as having incorrect bond stereo marks where previously only the left structure was identified.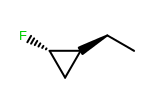
OEMol can now represent conformer coordinates in OEHalfFloat (16-bit),
double(64-bit), andlong double(>=64-bit) precision. These molecule types can be accessed through the corresponding constants in theOEMCMolTypenamespace.The following features were added to work in parallel with this new feature:
Reading and writing OEHalfFloat (16-bit) precision conformers to the
OEFormat.OEBfile format, including rotor-offset-compressed files.Reading and writing
double(64-bit) precision conformers to theOEFormat.OEBfile format, including rotor-offset-compressed files.Reading and writing
double(64-bit) precision conformers to theOEFormat.XYZfile format. By default, the XYZ writer will introspect the molecule type to determine how many significant digits to write.OEOFlavor.XYZ.SinglePrecisionandOEOFlavor.XYZ.DoublePrecisioncan be used to force either 5 or 18 significant digits.OEMolBase.SetCoords,OEMolBase.GetCoords, andOEMCMolBase.NewConfcan now take coordinates in OEHalfFloat andlong doubleprecision.OEConfBase.GetCoordsPtrsignature has been changed for better performance. Users should still prefer using OEConstCoords and OEMutableCoords.Conformer geometry operations will now preserve the precision of the input conformer coordinates.
Note
These alternative precision conformer implementations are not used by default. They require the user to explicitly specify a constant from the
OEMCMolTypenamespace to the OEMol constructor.Warning
Older versions of OEChem, and thus many previously released OpenEye applications will not be able to read
OEBfiles created from these alternative precision conformer molecules. Care should be taken to only pass these files to toolkits and applications that use at least the 2.0.2 version of OEChem. Note, this is not the default, the default is still to generate OEB files that can interoperate with older versions of OEChem.
Major bug fixes
OEMDLStereoFromBondStereoandOEMDLPerceiveBondStereowill now generate consistent atom stereo when a wedge or hash bond occurs between atoms with an angle less than 180 degrees as shown in the following figure. This will make OEChem consistent with the official Accelrys definition of stereo in this case. As well as ensuring that SMILES can be round-tripped through an OEChem generatedSDFfile that has had 2D coordinates automatically generated.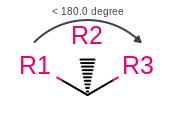
OEMDLHasIncorrectBondStereohad a major bug introduced in the 2014.Jun release. The bug was an incorrect array allocation and an out of bound access that resulted in nondeterministic processing of stereocenters and potential stack corruption. The 2014.Jun release should not be used for theOEMDLHasIncorrectBondStereofunction.OEWriteMoleculewill no longer improperly flip bond stereochemistry of complex macro-cycles when writing toOEFormat.MDL,OEFormat.SDF, orOEFormat.CDXfile formats. This was caused by the addition of automatically generating 2D coordinates when writing to these file formats in OEChem 2.0.0, the 2014.Feb release. In cases when the generated 2D coordinates are inconsistent with the cis/trans bond stereo configuration of the molecule, the coordinates will be zeroed out, i.e., the molecule will be written with no coordinates. This can occur when generating 2D coordinates for a molecule that contains a macro-cycle with specified cis/trans stereo bond configuration like the following: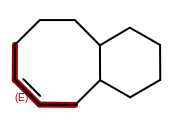
Writing to the
OEFormat.SDFandOEFormat.MDLfile formats will no longer improperly promote structures with relative stereochemistry to absolute stereochemistry in some cases. This case was when an input structure had atom stereomarks and a molecule chiral flag of ‘0’ (i.e. relative stereochemistry) erroneously causing a promotion of relative stereochemistry to absolute stereochemistry on export toOEFormat.SDForOEFormat.MDL. This change only affects structures that originate from the V2000 and V3000 molfile formats without 3D coordinates. Structures from SMILES and other formats don’t represent relative stereochemistry, therefore, those formats will still only translate absolute stereochemistry.OEMolDatabase.GetTitlewould sometimes return an empty title for multi-conformer OEB files that had their titles stored on the conformer.OEMolDatabase.GetTitlewill now recognize this case and use the title from the first conformer if the top-level OEMCMolBase does not have a title.Rotor-offset-compressed OEB files created on big-endian machines should now be readable on little-endian machines again. This bug was introduced in the 2011.1 release, the OEChem 1.7.5 release. The only big-endian architecture supported since then was PowerPC, and that support was dropped due to lack of usage.
OEGetDistance2that takes a OEConfBase will now return the square of the distance instead of the distance as it should. This bug was introduced in 2014.Feb, the OEChem 2.0.0 release. OEGetDistance2 functions that take arguments other than OEConfBase, e.g, OEMolBase, never had a bug introduced and will continue to return the square of the distance between the two atoms.OECreateInChIwill no longer crash and instead throw a warning when the InChI library returns an empty string.OEConfBase.GetTitlewill be preferred over theOEMolBase.GetTitlewhen writing multi-conformer OEMCMolBase molecules out to file formats that support titles. Previously, users were required to set the OEMCMolBase title to an empty string,mol.SetTitle(""), to achieve this behavior. This code will continue to work, but is no longer necessary to get the more specific conformer titles to be written out.OEBfiles will continue to write out both titles.OEConstCoords and OEMutableCoords will no longer crash when given an empty molecule.
Minor bug fixes
OE3DToAtomStereo, for the perception of atom stereo from 3D coordinates, depends on the following properties:OEPerceived.RingAtomsAndBonds,OEPerceived.Aromaticity, andOEPerceived.Chiral. If any of these properties are un-set when callingOE3DToAtomStereo, they will be automatically reperceived before calculating the atom stereo properties.OE3DToBondStereo, for the perception of bond stereo from 3D coordinates, depends on the following properties:OEPerceived.RingAtomsAndBonds,OEPerceived.Aromaticity, andOEPerceived.Chiral. If any of these properties are un-set when callingOE3DToBondStereo, they will be automatically reperceived before calculating the bond stereo properties.OEOFlavor.ISM.ExtBondswill no longer be used when generating SMILES withOEMolToSmiles. This will causeOEMolToSmilesto generate the same canonical isomeric SMILES as when writing into anOEFormat.ISMfile format.OEChem::OEMCMolType::TorsionandOEChem::OEMCMolType::Rotationconstants have been removed.OEInitHandlercalled on the options specified byOEBDefaultOptswill now initialize the handler to the exact same set of handlers asOEInitDefaultHandler. The noticeable difference between the two was thatOEInitHandlerwould inadvertently turn on writing out theOEBondBase.GetIntTypefield, resulting in larger file sizes.OEBondBase.GetIntTypewas also turned on for theOEBRotCompressOptsoptions as well.OEBondBase.GetIntTypewill now only be turned on by theOEBAtomIntTypeOptsoptions.OEMolBaseType.OEMiniMolwill now copy coordinates of other molecule implementations during copy construction.OEMCMolBase read through OEConfTestBase from non-SDF file formats should have a slightly smaller memory footprint. The
MDLParityflag was always being stored in generic data, regardless of whether the molecule had the property or not.OEWriteMoleculewill no longer change the active conformer of an OEMCMolBase while writing to file formats that do not support multi-conformer molecules like SDF and MOL2.OEWriteConstMoleculewill no longer ignore the flavors specified by the oemolostream for theOEFormat.XYZformat.Warning message about an invalid format used with
OEReadMoleculewas previously saying there was a problem withOEWriteMolecule. This was incorrect, and the message now states “OEReadMolecule” instead.Molecules read through
OEMolDatabase.GetMoleculewill no longer have their indices written to the OEB file format. This was just causing file size bloat and possibly confusing behavior withOEHasMolDBIdxwhen the molecule was written and then read again from OEB through an oemolistream.OEMolDatabase.GetTitlefixed for PDB files that include data beyond the 72nd column.Some redundant information is removed from OEB files written from OEMCMolBase objects. This should make OEB files the exact same size when round-tripping molecules now instead of becoming slightly larger.
OEFormat.SDFandOEFormat.MDLfile formats no longer support the long deprecated CPSS reaction fields. More deprecated atom and bond fields in the V2000 molfile format continue to be exported but will be removed entirely in future versions.OEMDLSetParityadded to allow explicitly setting the chiral flag status. This was added for symmetry withOEMDLGetParityand to allow explicit control of the exported chiral flag state by the user.SMILES containing explicit hydrogens in stereo configurations were being non-optimally depicted. SMILES parsing will now set the reaction role (
OEAtomBase.SetRxnRole) of stereo explicit hydrogens.OEMCMolBase.NewConfwill now return aNULLOEConfBase pointer instead of crashing when passed aNULLcoordinates pointer.
Documentation fixes
OEMolBaseTypeis now documented.OEOFlavor.XYZis now documented.All OpenEye code examples will now prefer to use
OEMolToSmilesandOESmilesToMolinstead of theOECreate*SmiStringandOEParseSmilesfunctions.
OEBio 2.0.2
Minor internal improvements.
OESystem 2.0.2
New features
Small performance improvements when writing generic data to OEB files.
OEGetHostEndianandOEGetHostEndianadded to make template programming generic binary readers and writers easier.OEGeom3DAllZeroadded to determine whether an array of floating point numbers is all zero.Many
OEMath::OEGeomfunctions can now automatically up-cast OEHalfFloat to float during their operation making working with OEHalfFloat more seamless and requiring less temporary arrays.OEGeom3DReflectadded to reflect coordinates around any given axis.OECopyArrayshould now be significantly faster forlong doubledata arrays.
Documentation fixes
OEBitVector.ToHexStringmore thoroughly explained.OEBitVector documentation table was malformed.
OEPlatform 2.0.2
New features
OEThread.Startnow returns a boolean value to indicate whether thread creation has failed.OEHalfFloat can now interconvert with
doubleandlong doublemore easily.
Minor bug fixes
OEFileRandomNamewill now throw a warning that it is deprecated and will be removed in a future release. It is very dangerous and prone to race conditions.OEMutex data members are no longer exposed as
publicaccess on Windows.
Documentation fixes
OEThread is now documented.
OEGrid 1.4.7
Minor internal improvements.