Tutorial: Predict hERG Toxicity of Drug-Like Molecules¶
OpenEye Model Building is a tool to build machine learning models that predict physical properties of small molecules.
In this tutorial, we will use the hERG toxicity floe to predict molecule toxicity as distinct classes of “High” or “Low.” The floe predicts the hERG toxicity of each molecule and explains the prediction based on chemical moiety. It also gives confidence intervals and other insightful information (listed at the end of this tutorial). The Floe Report provides a summary of the output.
Note: All other pretrained floes to predict molecule properties work the same way.
This tutorial uses the following floe:
hERG Toxicity Prediction for Small Molecules Using ML and Cheminfo Fingerprints
Create a Tutorial Project¶
Note
If you have already created a tutorial project, you can reuse the existing one.
Log into Orion and click the “Home” button at the top of the blue navigation bar on the left side of the Orion Interface. Then click on “Create New Project,” and in the pop-up window enter Tutorial for the name of the project and click “Save” button.
Orion home page¶
Floe Input¶
The input dataset contains several OERecord(s). The floe expects an OEMol from each record. These are the molecules for which the model will predict hERG toxicity. Note: uploading .csv, .sdf, and other common file formats to Orion should automatically convert them to datasets. Let this dataset be P_1.
Here is a sample record from the dataset:
OERecord (
Molecule(Chem.Mol) : c1ccc(c(c1)NC(=O)N)OC[C@H](CN2CCC3(CC2)Cc4cc(ccc4O3)Cl)O
)
There can be another string field containing hERG toxicity values (as “High” and “Low”) to validate against. The following dataset does not contain this field.
Input Data
Run the hERG Toxicity Floe¶
Click the “Floe” button on the navigation bar.
Click on the Floes tab.
Under Categories, select the OpenEye Model Building package.
A list of floes will now be visible to the right.
Launch the hERG Toxicity Prediction for Small Molecules Using ML and Cheminfo Fingerprints Floe, and a Job Form will pop up. Specify the following parameter settings in the Job Form.
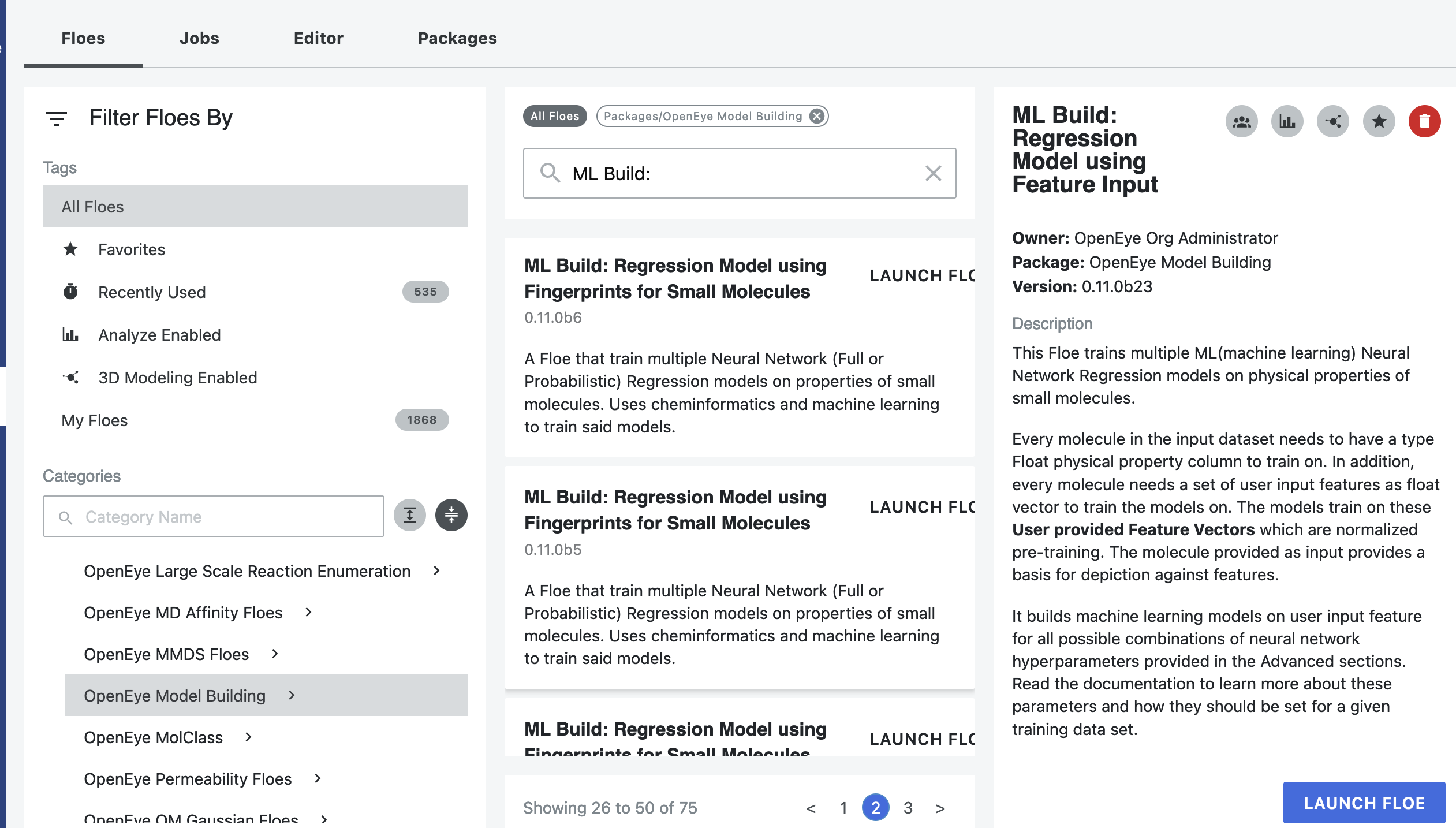
Choose P_1 as the Input Dataset.
The predicted molecules will be saved to the dataset listed in Output hERG Toxicity. Change the default name to something recognizable.

Click on the green “Launch Floe” button. That’s it! The floe will run, generate an output, and produce a Floe Report.
Analyze the OEModel Floe Report¶
The Floe Report should include information similar to what is shown below.
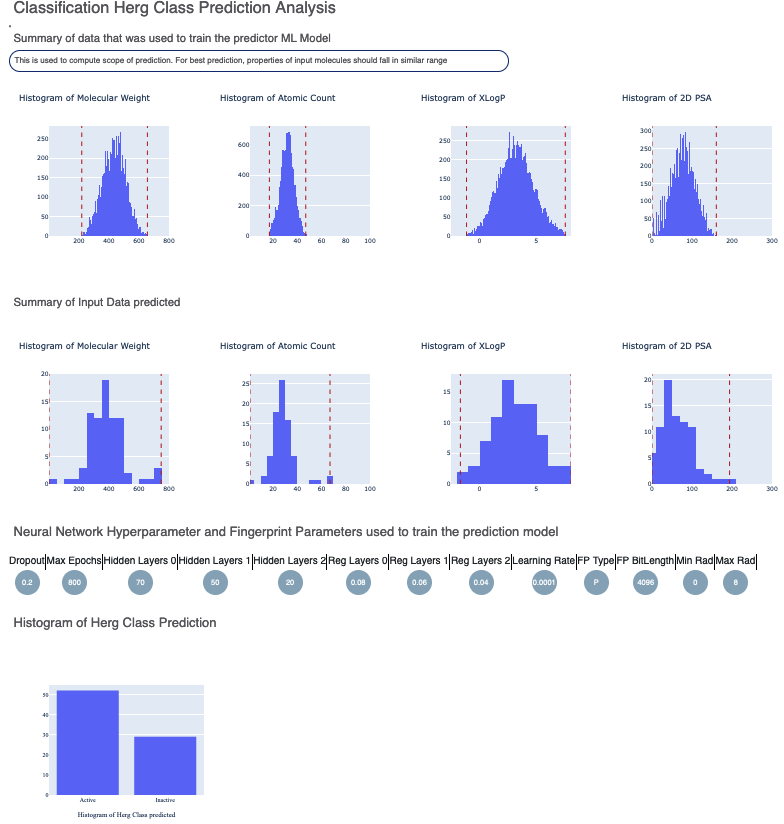
The top part contains the histogram summary of the data used to train the hERG ML model on. The red dotted lines illustrate the upper and lower quartile for the data. This is followed by the histogram summary of the input prediction data P_1. It is worthwhile to compare the x-axis ranges of the training and prediction data. If they are largely different, the prediction quality may deteriorate.
The next part contains the hyperparameter of the neural network the hERG model was trained on. Next is a histogram of the output prediction and the confidence with each prediction. We also have a plot for the confidence of prediction versus the actual output. These overall statistics help analyze the input molecules predicted.
Analyze the Output¶
Go to the Data page indicated on the navigation bar. A list of data produced by the floe will appear in the My Data folder.
This should have the same name you chose in the Output hERG Toxicity field. Activate the dataset by clicking on the circle with the plus sign next to the file name.
Navigate to the Analyze page on Orion; in the Spreadsheet Panel, you should see the molecules, their predicted pyrrolamide values, and the explanation of the output.
The output columns and their explanations are: * We assign ID numbers on each record molecule. This follows a linear ordering over all molecules. So if you activate both
the successful and failure predictions, and sort them based on number, the order should be the same as the input.
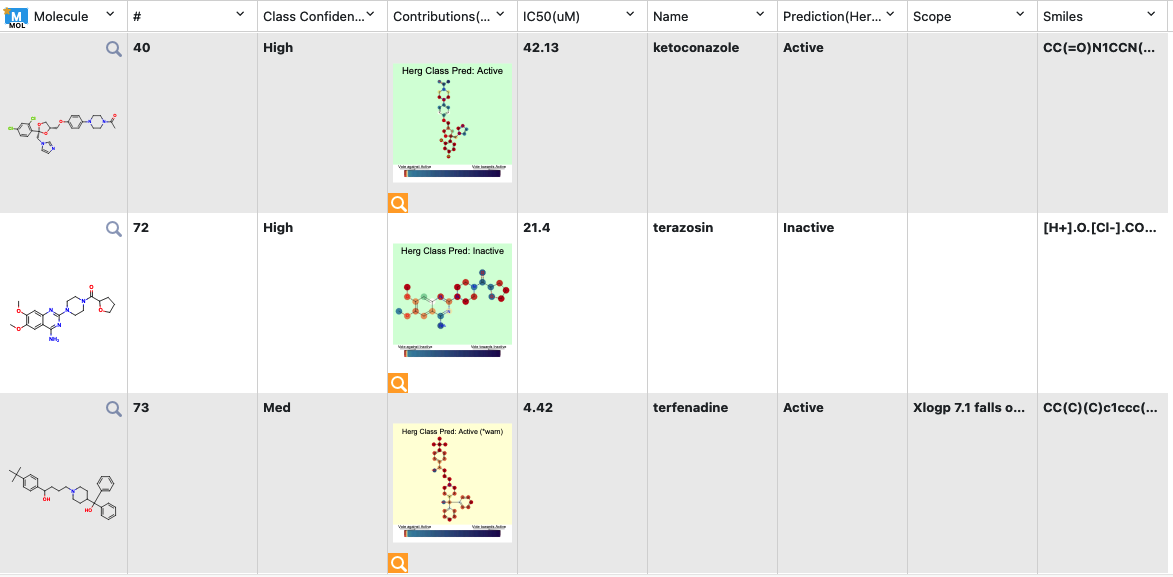
Class Confidence (hERG Toxicity): How confident the model is with its prediction as “High,” “Medium,” or “Low.” In this context, the prediction relates to the prediction confidence and not actual toxicity.
Contributions (hERG Toxicity): Explanation of prediction based on a local model. Based on the choice of molecule explainer (Atom by default), different parts will be color annotated with red denoting “vote against hERG active” while blue denotes the opposite.
Scope (8th column): The cause of an issue if there is an error or warning.
Predict (hERG Toxicity): Predicts toxicity as “High” or “Low.” The background color suggests how confident the model is, with green (most confidence), yellow (average confidence), and red (less confidence/out of scope). If the image has a red or yellow background, it means there is an error or warning issued as an additional column in the output (scope).