Tutorial: Building Machine Learning Regression Models on Feature Vector Input¶
OpenEye Model Building is a tool to build machine learning models that predict a physical property of small molecules.
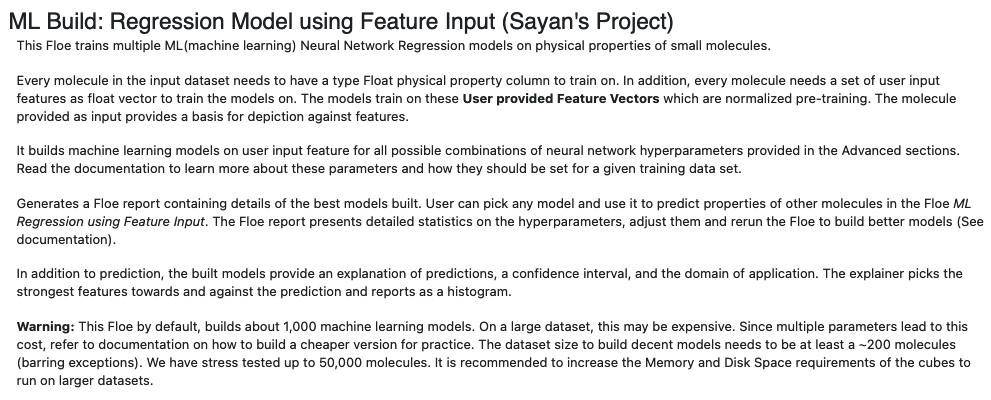
In this tutorial, we will train several regression models to predict IC50 concentrations at which molecules become reactant to pyrrolamides. The training data is known as the PyrrolamidesA dataset. The regression models will be built on user provided custom features and neural network hyperparameters. The results will be visualized in both the analyze page and Floe report. We will leverage the Floe report to analyze and choose a good model. Finally, we will choose a built model to predict the property of some unseen molecules.
This tutorial uses the following Floe:
ML Build: Regression Model using Feature Input
Warning
This tutorial keeps default parameters and builds around 1k machine learning models. While the per model cost is very cheap, based on the size of dataset, total cost might be expensive. For instance, with the pyrrolamides dataset (~1k molecules) it costs around $100. To understand the working of the Floe, we suggest building lesser models by referring to the Cheaper and Faster Version of the tutorial.
Create a Tutorial Project¶
Note
If you have already created a Tutorial project you can re-use the existing one.
Log into Orion and click the home button at the top of the blue ribbon on the left of the Orion Interface. Then click on the ‘Create New Project’ button and in the pop up window enter Tutorial for the name of the project and click ‘Save’.
Orion home page¶
Floe Input¶
The Floe requires an input dataset file, “Input Small Molecules to train machine learning models on”, with each record in the file an OEMolField .
There needs to be a separate field in this file containing the Float physical property to train the network on. This field needs to be selected in the promoted field parameter Response Value Field.
Finally, each record is required to contain a FloatVec feature set to train the network with. This field needs to be selected in the promoted field parameter Custom Feature. These features can be anything from molecule descriptors, to docking interactions, to electrostatic properties. All float vecs need to be of the same size. The idea is to allow scientists to generate their learnable set of parameters on which the models are to be trained. Thus, we are no longer bound by fingerprints, which though effective, have certain caveats.
The PyrrolamidesA dataset contains several OERecord (s). As stated, the Floe expects three things from each record:
An OEMol which is the molecule to train the models on
A Float value which contains the regression property to be learnt. For this example, it is the NegIC50 concentration.
A FloatVec value which contains the custom features on which the models will be trained. For this example, it is the floatVec field called “Feature”. For our example, this vector contains 11 properties of the molecule as PSA, XlogP etc.
Here is a sample record from the dataset:
OERecord (
*Molecule(Chem.Mol)* : c1cc(c(nc1)N2CCC(CC2)NC(=O)c3cc(c([nH]3)Cl)Cl)[N+](=O)[O-]
*NegIC50(Float)* : -2.6
*Feature(FloatVec)*: 25, 1.26, 2, 0.33, 4, 2, 1, 0, 0, 4, 104.15
)
Input Data
The input molecules and the training field values can be filtered and transformed by the preprocessing units:
Promoted parameter Negative Log : Applies -Log10 on training field values
Promoted parameter Preprocess Molecule :
Keeps the largest molecule if more than one is present in a record.
Sets pH value to neutral.
Promoted parameter Blockbuster Filter :
Applies the Blockbuster Filter
Run OEModel Building Floe¶
Click on the ‘Floe’ button in the left menu bar
Click on the ‘Floes’ tab
Under Categories select Packages
Select OpenEye Model Building
A list of Floes will now be visible.
Click on the ML Build: Regression Model using Feature Input and a Job Form will pop up. Specify the following parameter settings in the Job Form.
Click on the Input Small Molecules to Train Machine Learning Models on. button
Select the PyrrolamidesA dataset or your own dataset.
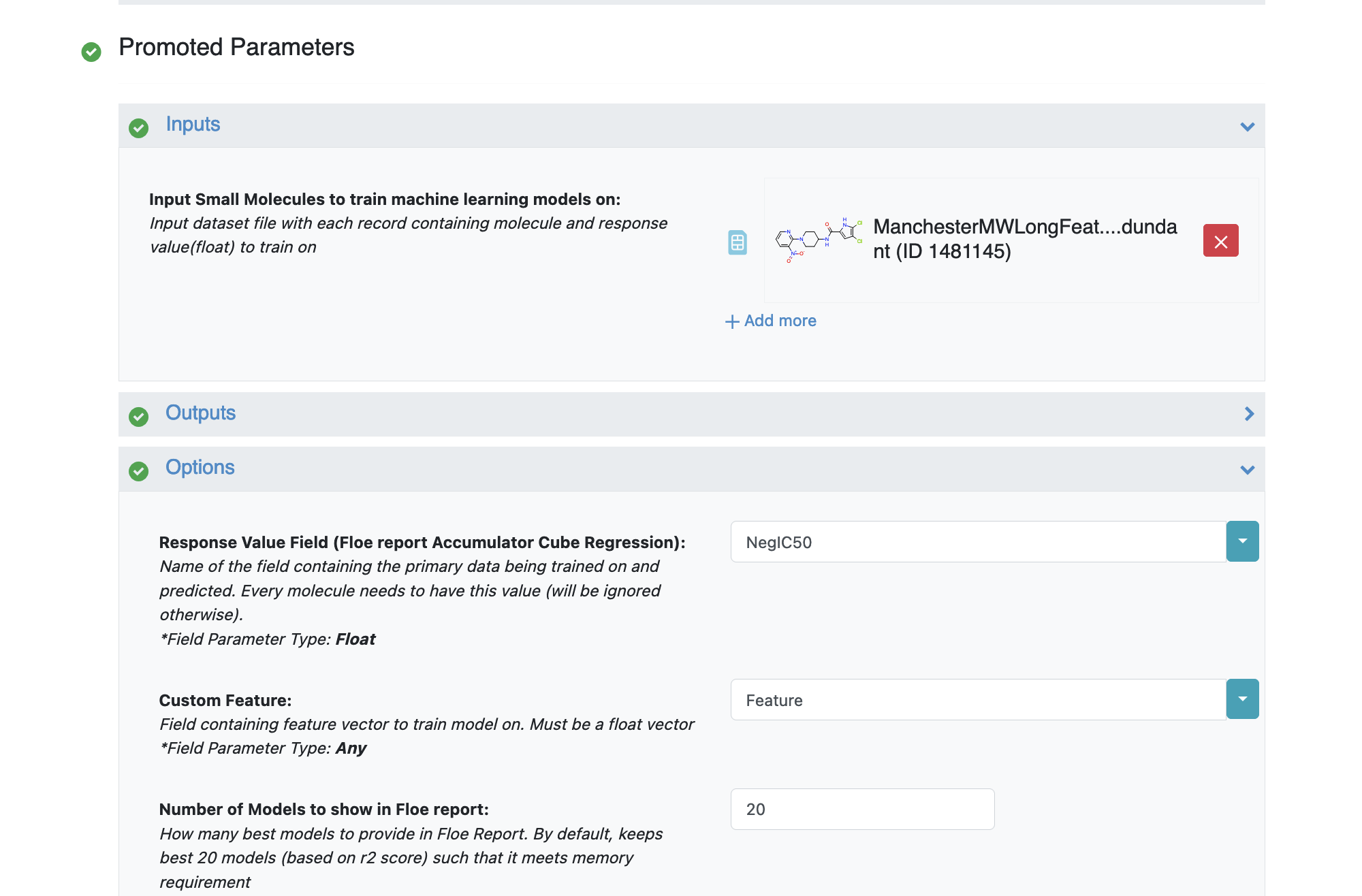
Under the Options tab, select the ‘Response Value Field’ which the model will train on. This field dynamically generates a list of columns to choose from based on the uploaded dataset’s columns. For our data, its NegIC50.
Under the same tab, select the Custom Feature which is the input training data fields for the model. For our case, choose ‘Feature’
Select how many model reports you want to see in the final Floe report. This field prevents memory blowup in case you generate >1k models. In such cases viewing the top 20-50 models should suffice.
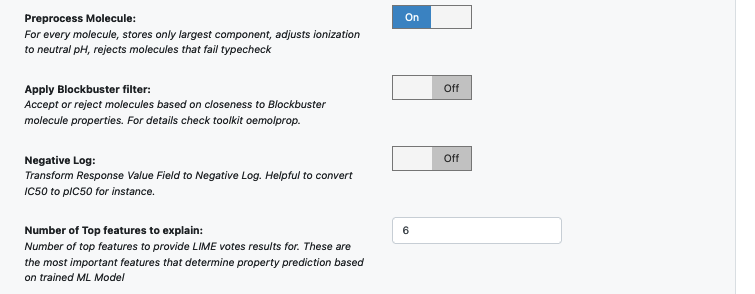
Next, turn on the Boolean parameter Preprocess Molecule. This preps every molecule by removing salts, charge, and multiple components, keeping the largest molecule. It also sets the ionization of the molecule to neutral pH.
Optionally, apply the blockbuster filter as well
Select how many ‘Number of Top features to explain’ you want to view for machine learning results prediction.
The model agnostic explainer lists which features from our input vector plays significant role in predicting the ‘Response Value field’. This parameter adjusts how many top features we want to see. Off course, the max length of this field should be <= the length of our input feature vector (11 in this case). We recommend keeping the number below 10 (show top 10 most significant feature votes) as the explainer diagrams get difficult to interpret if there is too much data.
In case it is necessary to transform the training values (Pyrrolamides in this case) to negative log, turn on the Negative Log switch as well. This is helpful when the data needs to be converted from IC50 to NegLogIC50 for instance. Keep it off for this example.
Change the default name of outputs to something recognizable.
You can let the model run at this point and it should run succesfully with default parameters. However, we can tweak a few parameters to learn more about the functionality.
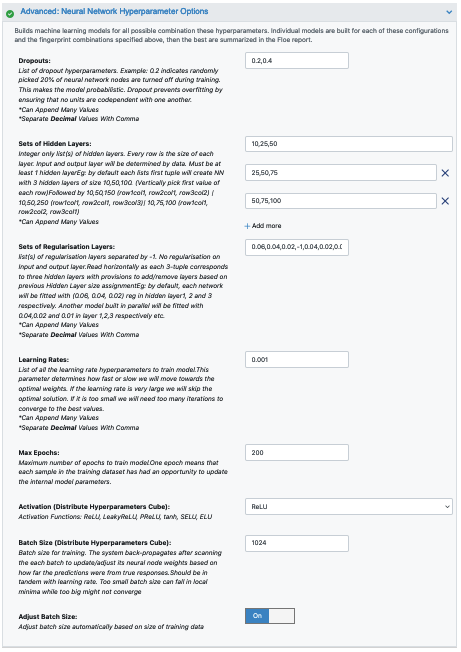
Next we move to the Advanced: Neural Network Hyperparameters Options. This is where the Machine Learning parameters are listed. Again, we can leave them as defaults, or choose to add/modify a few values based on the How-To-Guide.
We add Dropouts to prevent overfitting.
Next, let us inspect the parameter Sets of Hidden Layers.
Since it is a -1 separated list, by default there are 3-layer networks of size (250,150,50) and (150,100,80).
This gives a total node size of 330(150+100+80) and 240(100+80+60) in the default models.
Plugging in the formula stated in the How-to-Guide building Optimal Models, we should probably reduce the number of nodes to prevent overfitting.
Another important hyperparameter is the ‘Sets of regularisation Layers’. This field sets L2 regularisation for each network layer, making it a -1 separated 3-tuple list as well.
Increasing the Learning Rates is a way of speeding things up, although the algorithm may not detect the minima if this value is too high. We can train our model on multiple learning rate values. Leave defaults here.
If the dataset is big, we may consider increasing the Max Epochs size as it may take the algorithm longer to converge. Leave defaults here.
Activation: RelU is the most commonly used activation function for most models. Change to one of the options in the list if needed (see How-to-Guide on building Optimal Models).
The Batch Size defines the number of samples that will be propagated through the network. With larger batch sizes there is a significant degradation in the quality of the model, as measured by its ability to generalize. However, too small of a batch size may take the model a very long time to converge. For a dataset size of ~2k, 64 is probably a good batch size. However for datasets of saround 100k, we may want to increase batch size to at least 5k.
Set Batch size to 512
That’s it! Lets go ahead and run the Floe!
Note
The default memory requirement for each cube has been set to moderate to keep the price low. Although this floe is even cheaper than the fingerprint based floes, if you are training larger datasets, be mindful of the time it takes for the cubes to run into completion. Increasing the memory will let Orion assign instances with more processing power, thereby resulting in a faster completion. In some cases, the cubes may run out of memory (indicated in the log report) for large datasets. Increasing cube memory will fix this as well.
Analysis of Output and Floe Report¶
After the Floe has finished running, click the link on the ‘Floe Report’ tab in your window to preview the report. Since the report is big, it may take a while to load. Try refreshing or popping the report out to a new window if this is the case. All results reported in the Floe report are on the validation data. The top part summarizes statistics on the whole input data.
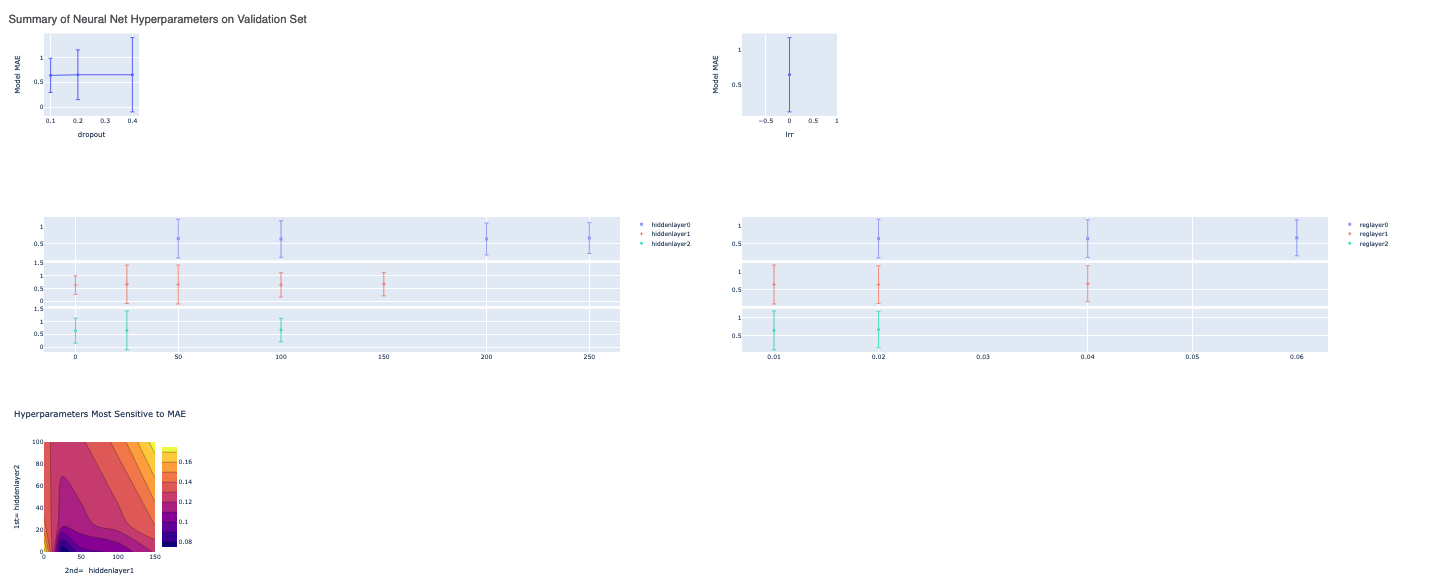
The graphs show the Mean Absolute Error (MAE) for different values of the neural network hyperparameters. It helps us analyze how sensitive the different hyperparameter values are and plan future model builds accordingly. There is also a plot between the top two most sensitive hyperparameters.
Next, we tabulate the list of all models built by the fully connected network. These models are sorted by the least R2 Score (for validation data). On the face of it, the first row should be the best model since it has the least error on the validation data. But there can be several other factors besides the best R2 to determine this, starting with Loss in the next column. Hence, lets look at a sample model by clicking the ‘Model Link’. This will take you to a new Floe report page.
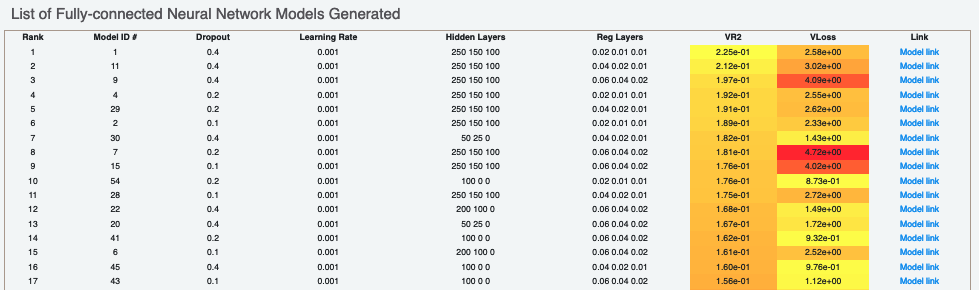
For each model, we have a linear color scale showing the rank. We also have the cheminformatics and machine learning parameters the model was trained on.
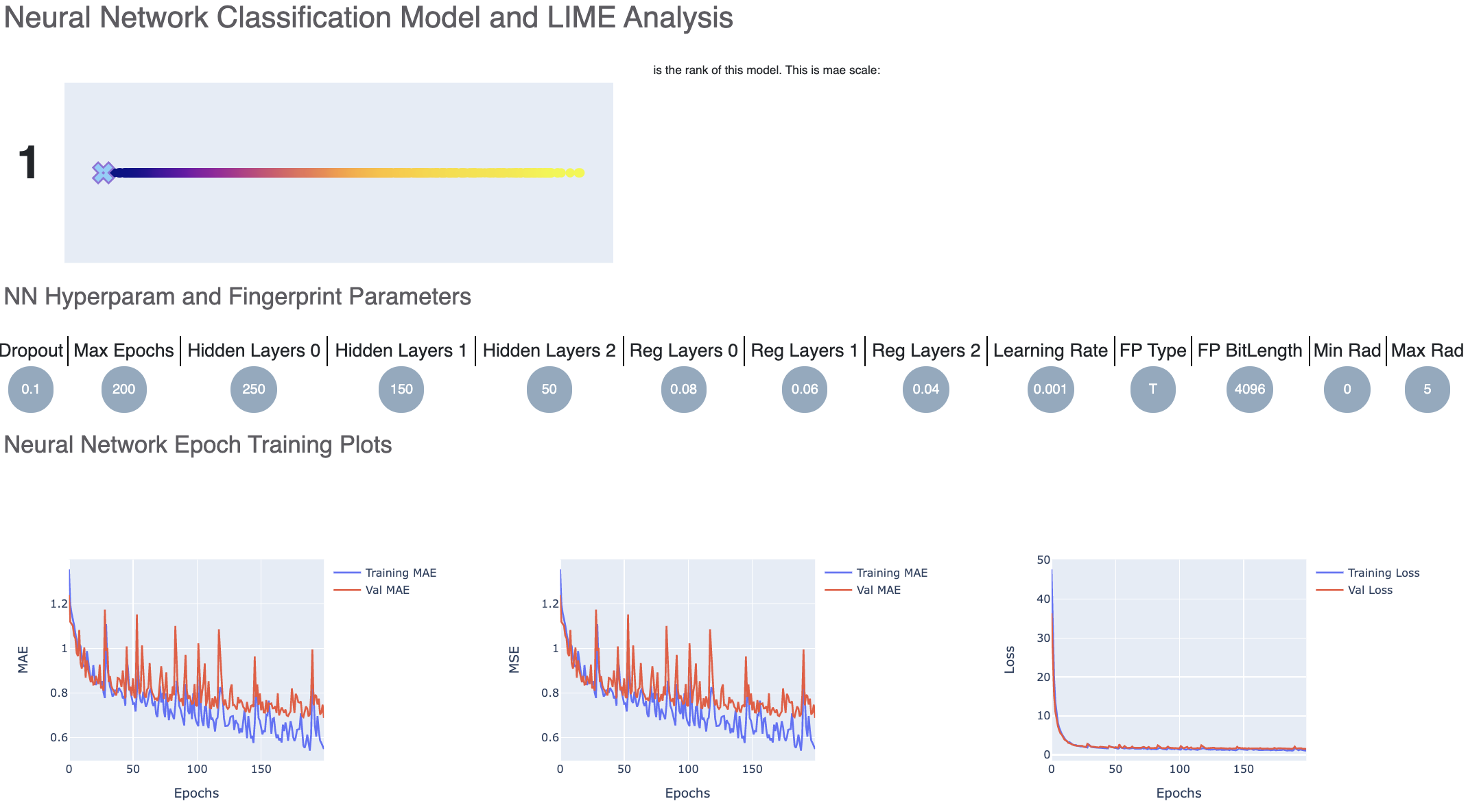
Let’s look at the training curves under ‘Neural Network Epoch Training Plots’. We see that the training and validation MAE follow a similar trend which is a good sign of not overfitting. Had they been diverging, we might have to go back and tweak parameters such as number of hidden layer nodes, dropouts, regularizers etc. If there were too much fluctuation in the graph it would suggest we need a slower learning rate.
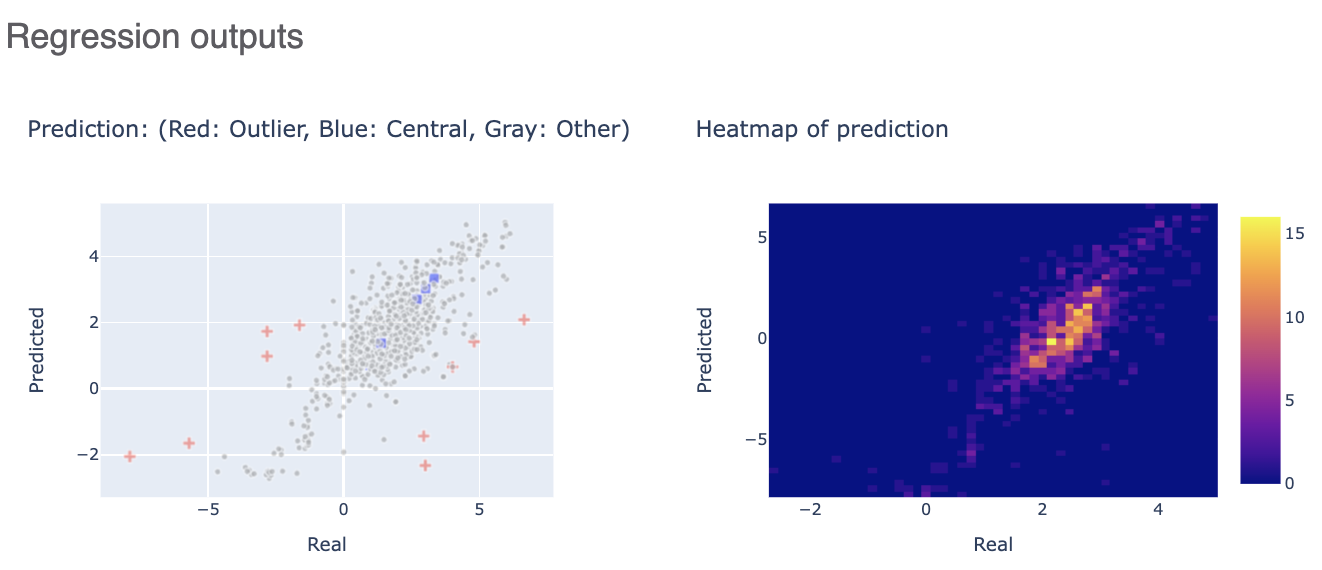
The Regression outputs plots real versus predicted to show how well the training is correlated.
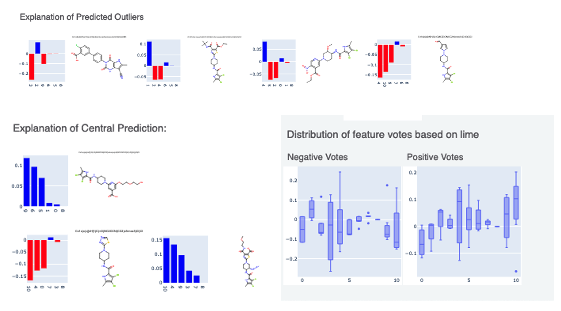
Finally, clicking on the link to interesting molecules takes us to another page which has some examplary Central and Outlier Molecule. Note that unlike fingerprints, we cannot map features back to molecules so our explainer now is a graph listing the top features which voted most strongly to the prediction. The x-axis count of this plot can be altered by the Number of feature promoted parameter we encountered earlier.
The floe report ends with a plot of the features which contributed either towards or against the response value trained on. This plot is very insightful as it gives an account of which features made the most contributions across all the molecules in the validation set.
While this tutorial shows how to build regression models, building an optimized model is a non-trivial task. Refer to the How-to-Guide on building Optimal Models.
After we have found a model choice, we can use it to predict and or validate against unseen molecules. Go to the next tutorial Use Pretrained Model to predict generic property of molecules to do this.