Tutorial: Use Custom Feature Input to predict regression properties¶
OpenEye Model Building is a tool to build machine learning models that predicts molecular properties.
In this tutorial, we will use a trained fully connected neural network model to predict physical property of molecules. The model was trained on a feature vector the user provided. This floe expects a feature vector of the same length as training set to make predictions on. The Floe predicts the property of each molecule and explans it. It also generate confidence intervals and other info. The Floe report provides a summary of the output.
This tutorial uses the following Floe:
ML Predict: Regression using Feature Input.
It is suggested to read the previous tutorial to learn how to build a machine learning model to use for prediction.
Create a Tutorial Project¶
Note
If you have already created a Tutorial project you can re-use the existing one.
Log into Orion and click the home button at the top of the blue ribbon on the left of the Orion Interface. Then click on the ‘Create New Project’ button and in the pop up window enter Tutorial for the name of the project and click ‘Save’.
Orion home page¶
Floe Input¶
The inputs required are:
Molecule Dataset (P_1)
Tensorflow Machine Learning Custom Feature Input Regression Model Dataset (M_1)
The P_1 dataset contains several OERecord (s). The Floe expects an OEMol from each record. This is the molecules the model will predict physical property of. Let this dataset be P_1. Corresponding to an OEMol, the dataset also needs to contain a float vec field for every record and the length of this float vec need to be the same. For csv files, you can convert comma separated float values in a single column to such floatvecs. Orion automatically converts csvs to datarecords ones uploaded.
Here is a sample record from the dataset:
OERecord (
*Molecule(Chem.Mol)* : c1cc(c(nc1)N2CCC(CC2)NC(=O)c3cc(c([nH]3)Cl)Cl)[N+](=O)[O-]
*Feature(FloatVec)*: 25, 1.26, 2, 0.33, 4, 2, 1, 0, 0, 4, 104.15
)
There can be another float field containing the physical property values to validate against. The following dataset does not contain said field.
Input Data
The M_1 dataset contains one or more machine learning models. To learn how to generate these models, read the previous tutorial on building models in Orion. We attach a sample trained Tensorflow Model to predict IC50.
Input Machine Learning Models
Run Generic Property Prediction Floe¶
Click on the ‘Floes’ button in the left menu bar
Click on the ‘Floes’ tab
Under the ‘Categories’ tab select ‘OpenEye Model Building’ package
In the search bar enter ML Predict
A list of Floes will now be visible to the right
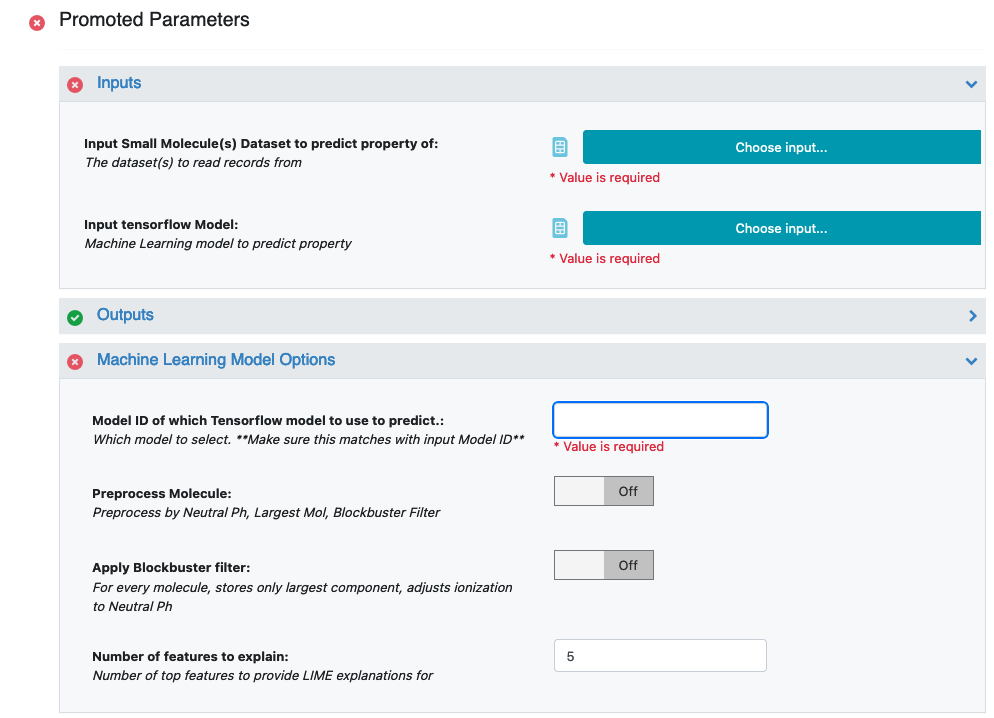
Launch the floe ML Predict: Regression using Feature Input and a Job Form will pop up. Specify the following parameter settings in the Job Form.
For the Input Tensorflow Model, choose M_1 from above. Make sure you have the ID of “Model ID of which Tensorflow Model to Use to Predict” to 1.
For the Input Small Molecule Dataset, choose P_1 from above
For the Custom Feature field, choose Feature. This is the feature vector that the ML Model was trained on, and the input on which property for P_1 will be predicted.
All the molecules predicted will be saved to name in the field Output Property. Change it to something recognizable.
Use the Model ID of which Tensorflow model to use to predict. field to identify the model ID you would like to predict the properties of your molecules. You can go to the analyze page on the model dataset to figure out the model ID of which you would want the predictions to be based on. More information to select this model is in the Howto-Guide: Use built machine learning model for property prediction/verification of unseen molecules.
Additionally if your small molecule dataset has a field containing the predicted value (sourced from elsewhere), you can turn the validation option on to get a statistical comparison between this value and the prediction.
We recommend the Preprocess Molecule to be same as what was used for training.
- Finally, the field Number of features to explain to how many top features to be explained
The model agnostic explainer lists which features from our input vector plays significant role in predicting the ‘Response Value field’. This parameter adjusts how many top features we want to see. Off course, the max length of this field should be <= the length of our input feature vector (11 in this case). We recommend keeping the number below 10 (show top 10 most significant feature votes) as the explainer diagrams get difficult to interpret if there is too much data.
That’s it ! Things should run, generate an output and a Floe report
Analyze OEModel Floe Report¶
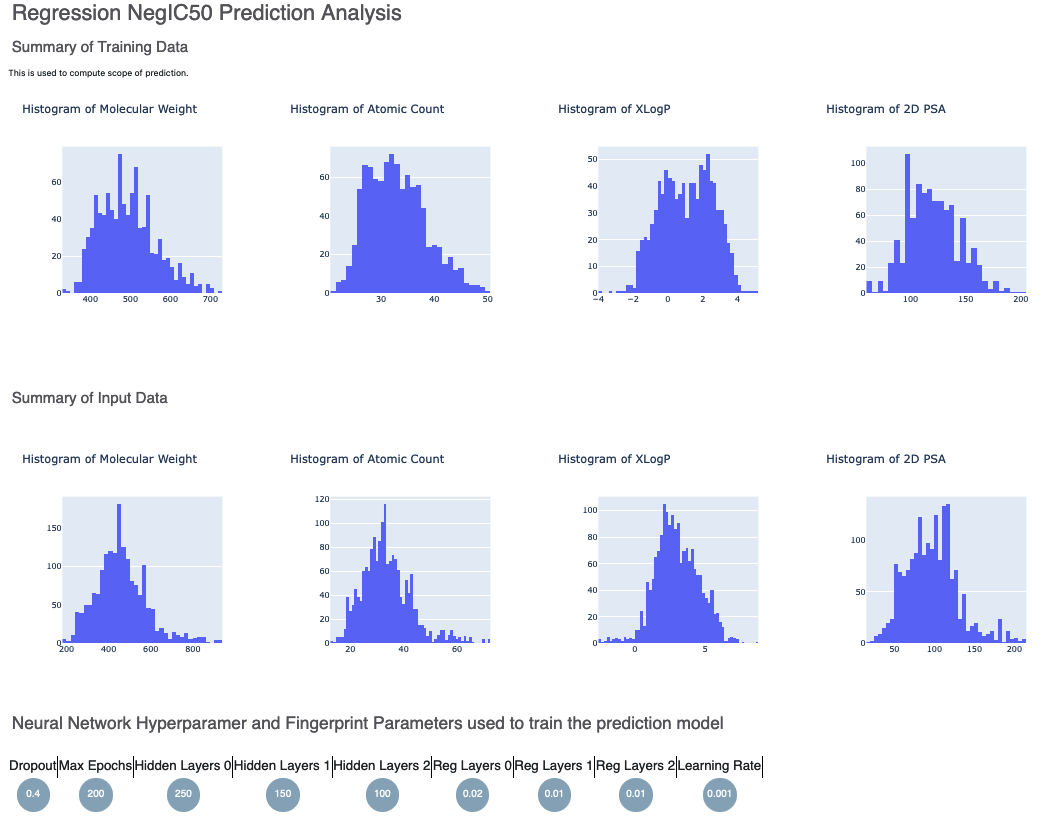
Here is a sample image of how the Floe report should look (Assuming you ran a fully connected neural network model like M_1):
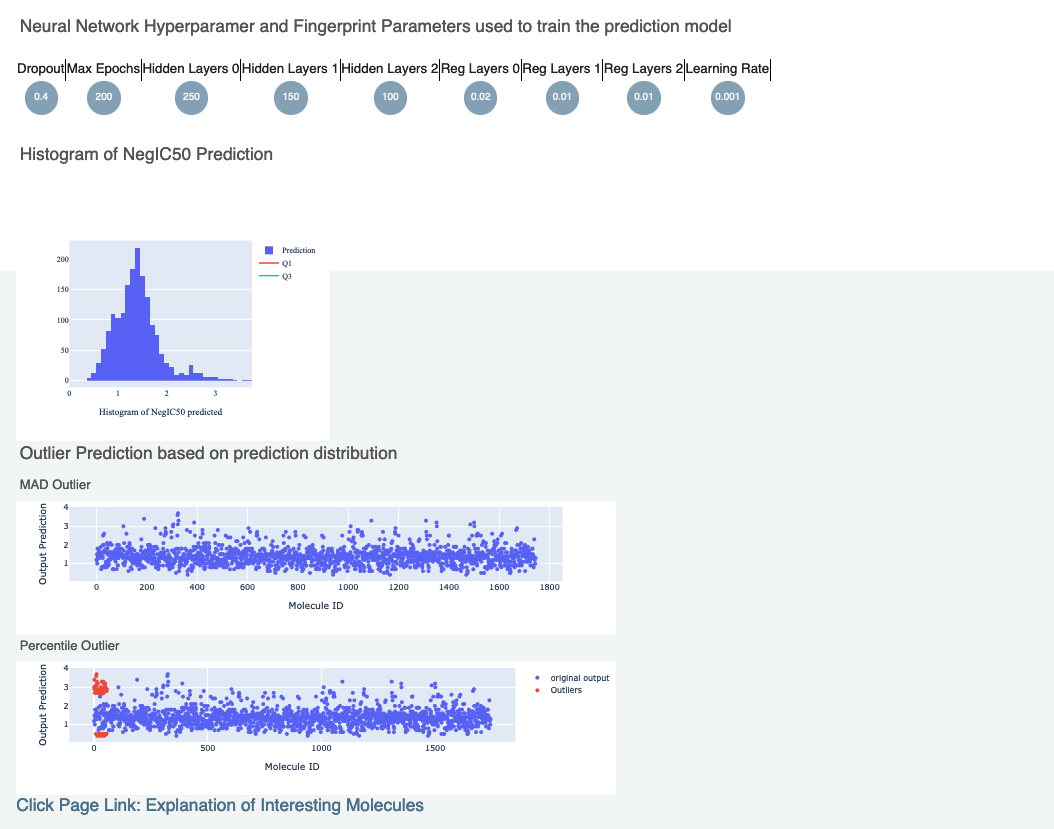
The top part contains histograms of the training data and the data the floe was ran on. This is followed by the histogram of the output prediction. Finally we have parameters the model was trained on. These overall statistics help analyze the input molecules predicted.
We also have outlier prediction using:
MAD Technique: Median of Absolute Deviation
Percentile Technique: Percentile Outlier
Lastly, there is a link to a page under Interesting Molecules and it shows the annotated images that explain the outlier and central molecules.
Analyze Output¶
Go to the data section of Orion and Activate the data the Floe produced. This should have the same name you chose for the Output Prediction field of your Floe. The data can be activated by clicking on the small plus sign in a circle right next to it.
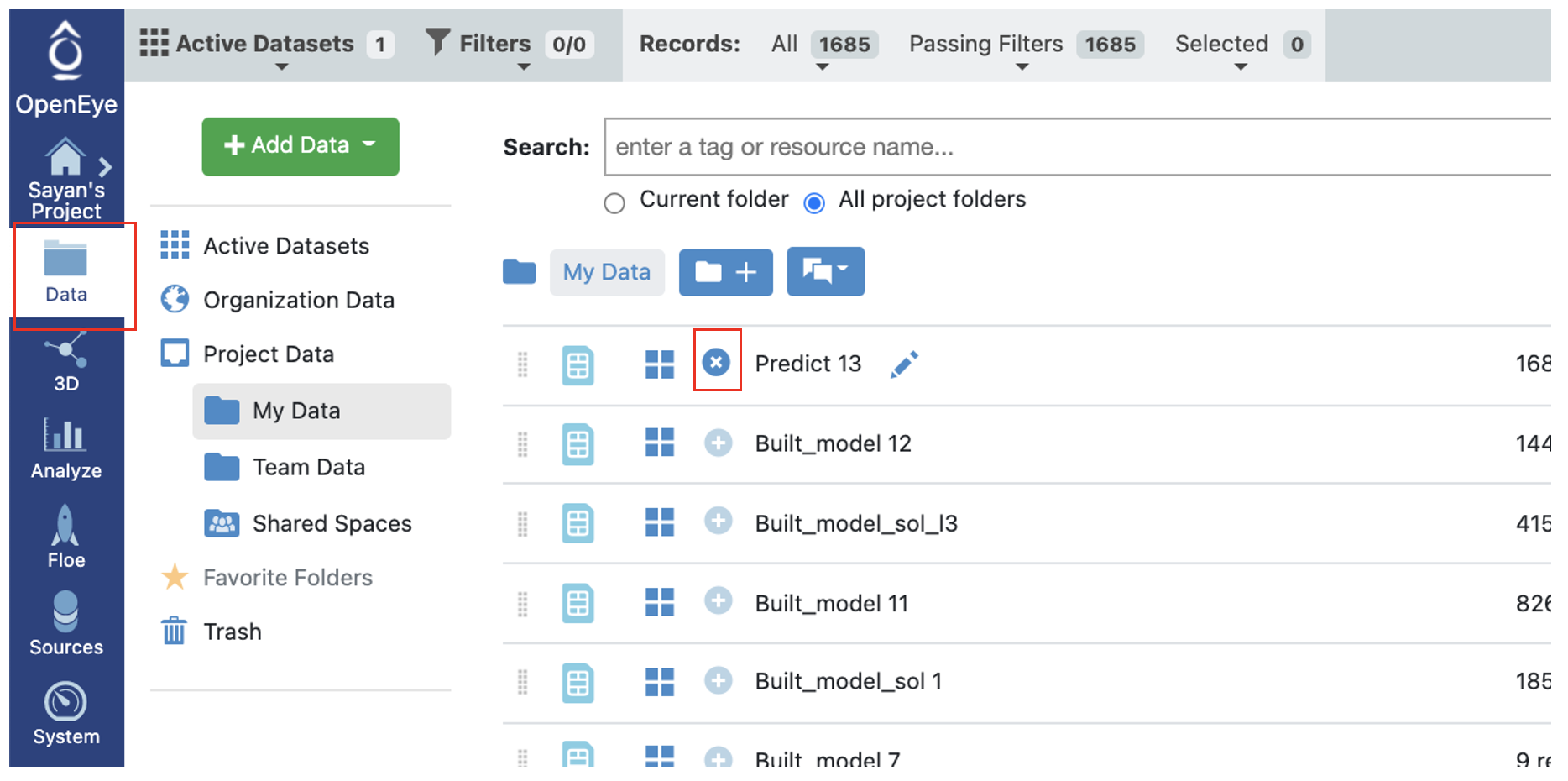
Now going to the analyze page in Orion, you should be able to see the molecules, their predicted pyrrolamide values, and the explanation of the output. Note that unlike fingerprints, we cannot map features back to molecules so our explainer now is a graph listing the top features which voted most strongly to the prediction. The x-axis count of this plot can be altered by the Number of feature promoted parameter we encountered earlier.
There are three output ports: Success, Failure, and No Confidence.
Outputs Meaning
Failure Data: (a) Molecule is too large or too small. or, (b) Molecule has an unknown atom.
No Confidence Data: Molecule’s property falls out of scope of training set. In this case, the model predicts with no guarantees. Explainer image has a red background.
Success Data: (a) Falls within scope; explainer has green background. (b) Falls at the edge of scope; explainer has yellow background.
The output columns and their explanations are:
Class Confidence(Property Unit): High, Med or Low confidence in prediction.
Class Prediction(Property Unit): High, Med or Low prediction of property. Depends on if the value lies within <25% (Low), >75%(High), or in between.
Contributions: Explanation of prediction based on a local model. Unlike fingerprints, we cannot map features back to molecules so our explainer now is a graph listing the top features which voted most strongly to the prediction. The x-axis count of this plot can be altered by the Number of feature promoted parameter we encountered earlier.
Scope: If there is an error or warning, what caused the issue.
Prediction(Property Unit): Physical property prediction of the molecule.
Note
We assign IDs (#) on each record molecule. This follows a linear ordering over all molecules. So if you activate all the successful, no confidence, and failure predictions, and sort them based on #, the order should be same as input.