Grapheme TK 1.3.0
New features
Due to the deprecation of OEFragmentNetwork and related classes in OEBio, Grapheme TK has introduced the following overloads:
OEPrepareActiveSiteDepictionfunction has been overloaded. The new version takes an OEInteractionHintContainer object.OE2DActiveSiteDisplay class now should be constructed with an OEInteractionHintContainer object.
All functions and methods using OEFragmentNetwork are deprecated in Grapheme TK and will throw a warning when called. Active site depictions generated with the deprecated API will have a “deprecated” watermark (see the example below).
using the deprecated OEFragmentNetwork class
using the new OEInteractionHintContainer class
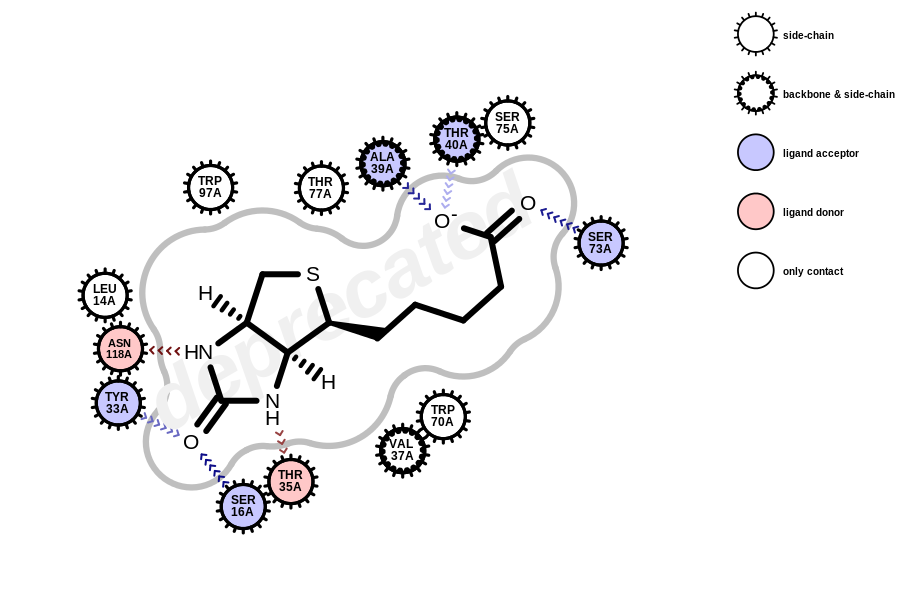
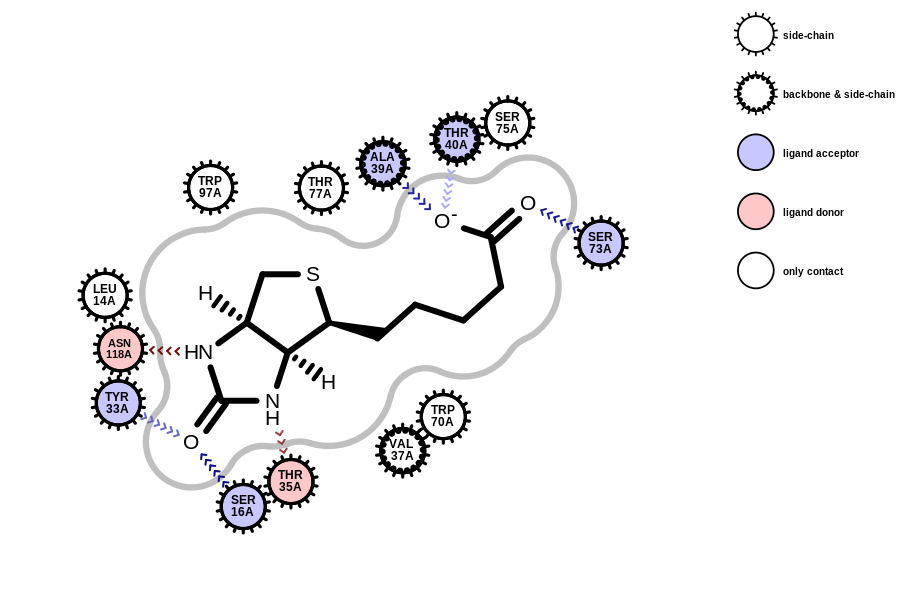
In order to avoid the “deprecated” watermark from appearing in the image, use
OEInteractionHintContainer asite(protein, ligand);
instead of
OEFragmentNetwork asite(protein, ligand);
See also
For more details about the deprecated classes and functions, see the Deprecated OEFragmentNetwork and related API section.
OE2DActiveSiteDisplay now can visualize salt-bridge and aromatic ring stacking interactions.
The color scheme used to visualize different interaction types when depicting protein-ligand interaction has been changed to accommodate the new types (Pi- and T-stacking and salt-bridge).
2016.Feb color scheme
2016.Jun color scheme
OE2DActiveSiteDisplay::GetDisplayedLigandandOE2DActiveSiteDisplay::GetDisplayedResiduesmethods have been added.OEAddLigandHighlightingandOEAddResidueHighlightingfunctions have been added to allow highlighting in active site depiction. See the example below.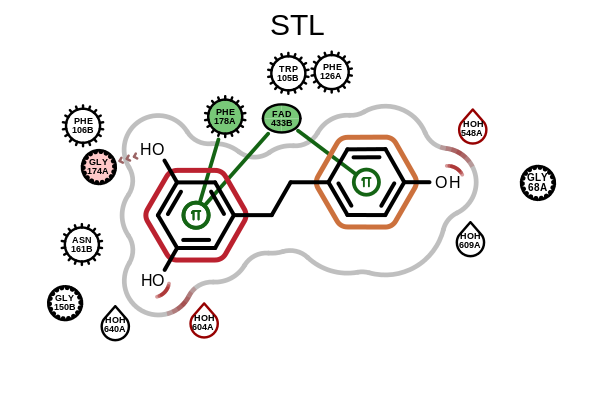
Example of ligand highlighting
OEShapeOverlapDisplayStyle::PropertyCloudstyle has been added to visualize shape overlays along with the following APIs:OEShapeOverlapDisplayStylenamespaceOEShapeOverlapDisplayOptions::SetOverlapDisplayStyleandOEShapeOverlapDisplayOptions::SetOverlapDisplayStylemethods
Example of visualizing shape overlaps with PropertyCloud style
Using this new style can result in a 10-times performance increase when generating shape overlay images; the generated images will also be significantly smaller (PNG 50%, SVG 60-times, PDF 9-times).
The following APIs have been added to identify ligand atoms and bonds, along with residues of an
OE2DActiveSiteDisplayobject that are close to a given 2D display coordinate:OEGetNearestAtomandOEGetNearbyAtomfunctionsOEGetNearestBondandOEGetNearbyBondfunctionsOEGetNearestResidueandOEGetNearbyResiduefunctions along with the OENearestResidue class
Documentation changes
All images in the Grapheme TK documentation were automatically regenerated to reflect the changes made since the previous release.
The example in Depicting Active Site Interactions has been updated to use the new OEInteractionHintContainer class instead of the deprecated OEFragmentNetwork.