Molecule Fragmentation
The OEMedChem TK currently provides four ways to partition a molecule into fragments:
OEGetRingChainFragments- fragments a molecule into ring and chain components.OEGetRingLinkerSideChainFragments- fragments a molecule into ring, linker and side-chain components as defined in [Bemis-1996] .OEGetFuncGroupFragments- fragments a molecule into ring and functional group components.OEGetBemisMurcko- fragments a molecule into ring, linker, framework and functional group components as in [Bemis-1996] .
These functions return an iterator over OEAtomBondSet objects that store the atoms and the bonds of the fragments.
The following examples (Listing 1, Listing 2) show how to
fragment a molecule into ring and chain components.
The code loops over the OEAtomBondSet objects
returned by the OEGetRingChainFragments.
Each OEAtomBondSet object is used to initialize an atom
and a bond predicates. These predicates specify which atoms and bonds have to be
considered when creating a subset of the molecule, i.e the fragment, when
calling the OESubsetMol function.
See the depiction of the input molecule and the generated fragments
in Figure: Example of fragmentation.
Listing 1: Example of molecule fragmentation.
from openeye import oechem
from openeye import oemedchem
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "COc1ccc(cc1)CC(=O)N")
for frag in oemedchem.OEGetRingChainFragments(mol):
fragatompred = oechem.OEIsAtomMember(frag.GetAtoms())
fragbondpred = oechem.OEIsBondMember(frag.GetBonds())
fragment = oechem.OEGraphMol()
adjustHCount = True
oechem.OESubsetMol(fragment, mol, fragatompred, fragbondpred, adjustHCount)
print(oechem.OEMolToSmiles(fragment))
The output of Listing 1 is the following:
CO
c1ccccc1
CC(=O)N
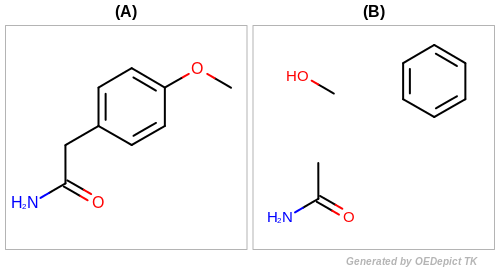
Example of fragmentation (A) input molecule (B) fragments returned by the OEGetRingChainFragments function
See also
OEAtomBondSet class and
OESubsetMolfunction in the OEChem TK manualPredicate Functors chapter in the OEChem TK manual.
The following example (Listing 2) shows how to
fragment a molecule into ring and chain components with annotations.
See the depiction of the input molecule and the generated fragments
in Figure: Example of Bemis Murcko fragmentation.
Listing 2: Example of molecule fragmentation with annotations.
from openeye import oechem
from openeye import oemedchem
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "CCOc1ccc(cc1)CC(OC)c2ccccc2CC(=O)N")
adjustHCount = True
for frag in oemedchem.OEGetBemisMurcko(mol):
fragment = oechem.OEGraphMol()
oechem.OESubsetMol(fragment, mol, frag, adjustHCount)
print(".".join(r.GetName() for r in frag.GetRoles()), oechem.OEMolToSmiles(fragment))
The output of Listing 2 is the following:
Framework c1ccc(cc1)CCc2ccccc2
Ring c1ccccc1.c1ccccc1
Linker CC
Sidechain CCO.CC(=O)N.CO
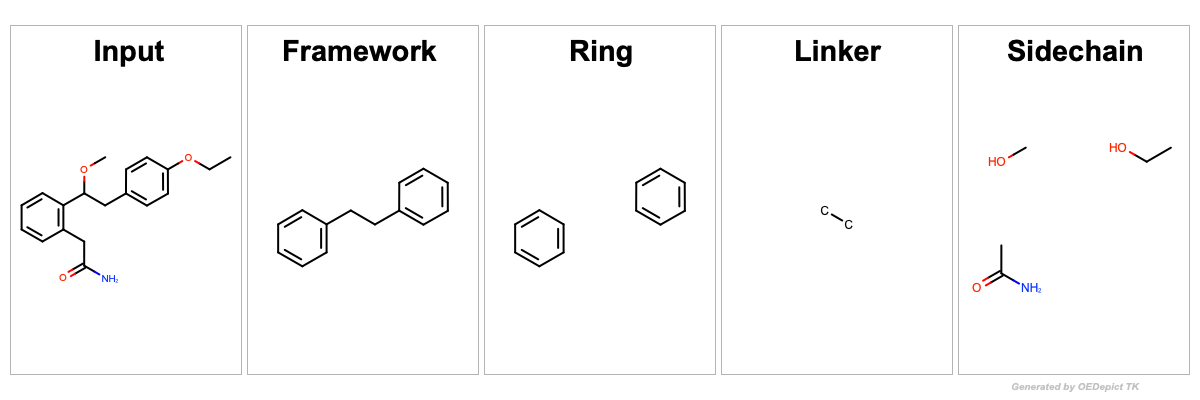
Example of Bemis Murcko fragmentation
The following example (Listing 3) shows how to
fragment a molecule and include unsaturated hetero bonds on the main framework.
See the depiction of the input molecule and the generated fragments
in Figure: Example of custom Bemis Murcko fragmentation including heteroatoms.
Listing 3: Example of custom molecule fragmentation.
from openeye import oechem
from openeye import oemedchem
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "CCc1nc(nc(n1)OC)NC(=O)NS(=O)(=O)c2ccccc2OC")
options = oemedchem.OEBemisMurckoOptions()
options.SetUnsaturatedHeteroBonds(True)
adjustHCount = True
for frag in oemedchem.OEGetBemisMurcko(mol, options):
fragment = oechem.OEGraphMol()
oechem.OESubsetMol(fragment, mol, frag, adjustHCount)
print(".".join(r.GetName() for r in frag.GetRoles()), oechem.OEMolToSmiles(fragment))
The output of Listing 3 is the following:
Framework c1ccc(cc1)S(=O)(=O)NC(=O)Nc2ncncn2
Ring c1ccccc1.c1ncncn1
Linker C(=O)(N)NS(=O)=O
Sidechain CC.CO.CO
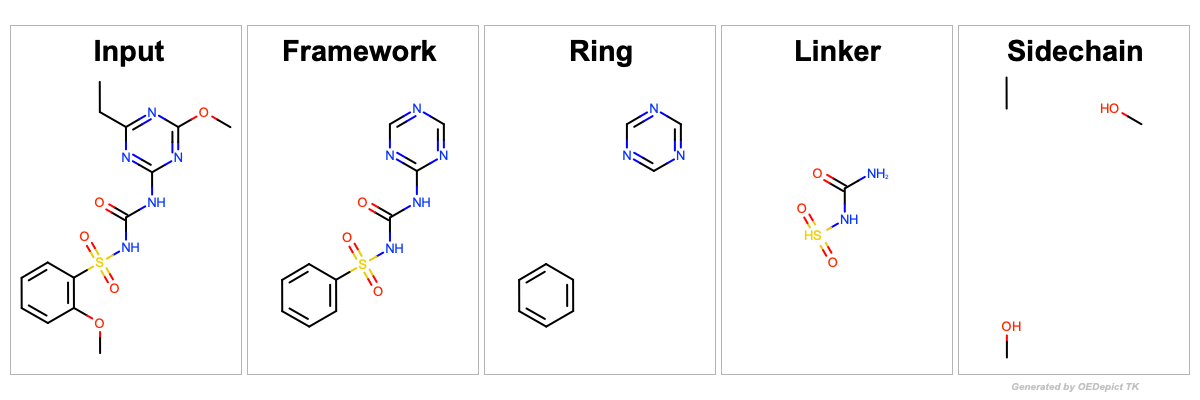
Example of Bemis Murcko fragmentation including unsaturated hetero bonds on main framework.
The following example (Listing 4) shows how to
fragment a molecule and include custom sidechains on the main framework.
See the depiction of the input molecule and the generated fragments
in Figure: Example of custom Bemis Murcko fragmentation including custom substituents.
Listing 4: Example of custom molecule fragmentation
from openeye import oechem
from openeye import oemedchem
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "CCc1nc(nc(n1)OC)NC(CC(=O)N)NS(=O)(=O)c2ccccc2CC(=O)N")
subsearch = oechem.OESubSearch()
subsearch.Init("[#6]-CC(=O)N")
options = oemedchem.OEBemisMurckoOptions()
options.SetSubstituentSearch(subsearch)
adjustHCount = True
for frag in oemedchem.OEGetBemisMurcko(mol, options):
fragment = oechem.OEGraphMol()
oechem.OESubsetMol(fragment, mol, frag, adjustHCount)
print(".".join(r.GetName() for r in frag.GetRoles()), oechem.OEMolToSmiles(fragment))
The output of Listing 4 is the following:
Framework c1ccc(c(c1)CC(=O)N)[SH4]NC(CC(=O)N)Nc2ncncn2
Ring c1ccc(cc1)CC(=O)N.c1ncncn1
Linker C(C(N)N[SH5])C(=O)N
Sidechain CC.CO.O.O
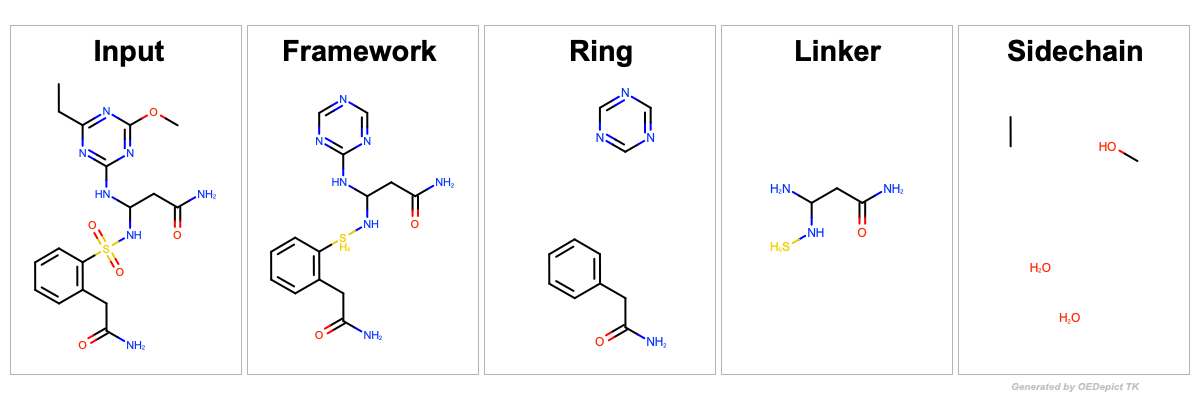
Example of Bemis Murcko fragmentation including custom substituents on framework