OEPrepareAlignedDepictionFrom3D
Link |
Description |
|---|---|
OEPrepareAlignedDepictionFrom3D(fitmol2D, fitmol3D, refmol2D, refmol3D) |
2D molecule alignment based on 3D |
OEPrepareAlignedDepictionFrom3D(fitmol2D, fitmol3D,, refmol2D, refmol3D, opts) |
2D molecule alignment based on 3D with options |
OEPrepareAlignedDepictionFrom3D(fitmol2D, fitmol3D, shapequery) |
2D molecule alignment to 3D shape query |
bool OEPrepareAlignedDepictionFrom3D(OEChem::OEMolBase &fitmol2D,
const OEChem::OEMolBase &fitmol3D,
const OEChem::OEMolBase &refmol2D,
const OEChem::OEMolBase &refmol3D,
bool clearCoords=true, bool suppressH=true)
Generates the 2D coordinates of a molecule driven by the 3D overlay of the 3D reference and the 3D fit molecules and the orientation of the 2D reference molecule.
- fitmol2D
The molecule for which 2D coordinates are generated.
- refmol2D
The 2D molecule whose orientation drives the 2D coordinate generation.
- fitmol3D, refmol3D
The 3D molecule whose overlay drives the 2D coordinate generation.
- clearCoords
If false and the fitmol2D has 2D coordinates, then these coordinates are used for the alignment. Otherwise the 2D coordinates of the fitted molecule are generated by calling the
OEPrepareDepictionFrom3Dfunction.- suppressH
If true, then explicit hydrogens are suppressed when generating the 2D coordinates of the fitted molecule. Only hydrogens that are necessary to faithfully represent tetrahedral stereochemistry will be kept.
bool OEPrepareAlignedDepictionFrom3D(OEChem::OEMolBase &fitmol2D,
const OEChem::OEMolBase &fitmol3D,
const OEChem::OEMolBase &refmol2D,
const OEChem::OEMolBase &refmol3D,
const OEAlignedDepictionFrom3DOptions &opts)
Generates the 2D coordinates of a molecule driven by the 3D overlay of the 3D reference and the 3D fit molecules and the orientation of the 2D reference molecule.
- fitmol2D
The molecule for which 2D coordinates are generated.
- refmol2D
The 2D molecule whose orientation drives the 2D coordinate generation.
- fitmol3D, refmol3D
The 3D molecules whose overlay drives the 2D coordinate generation.
- opts
The OEAlignedDepictionFrom3DOptions object that stores options controlling the generation of 2D coordinates.
The following code snippet shows how to use the OEPrepareAlignedDepictionFrom3D
function.
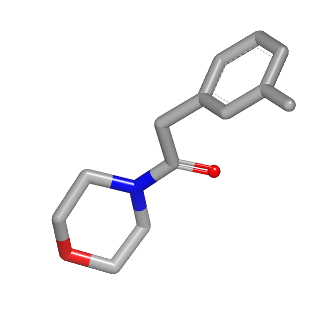
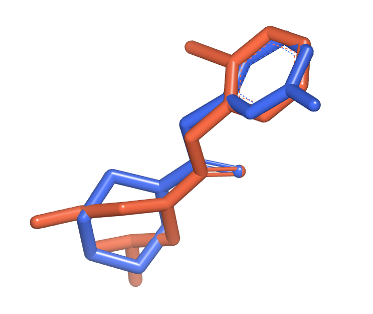
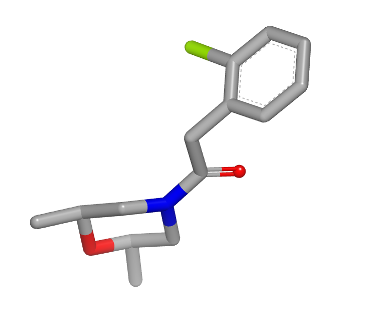
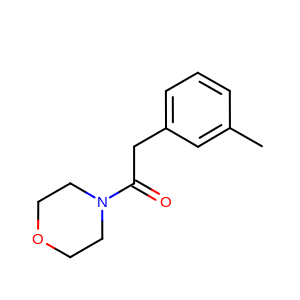
See generated images in Table: 2D depiction based on 3D molecule overlay.
refmol2D = oechem.OEGraphMol(refmol3D)
oegrapheme.OEPrepareDepictionFrom3D(refmol2D)
fitmol2D = oechem.OEGraphMol(fitmol3D)
oegrapheme.OEPrepareAlignedDepictionFrom3D(fitmol2D, fitmol3D, refmol2D, refmol3D)
width, height = 300, 300
opts = oedepict.OE2DMolDisplayOptions(width, height, oedepict.OEScale_AutoScale)
refscale = oedepict.OEGetMoleculeScale(refmol2D, opts)
fitscale = oedepict.OEGetMoleculeScale(fitmol2D, opts)
opts.SetScale(min(refscale, fitscale))
refdisp = oedepict.OE2DMolDisplay(refmol2D, opts)
oedepict.OERenderMolecule("OEPrepareAlignedDepictionFrom3D-ref-2D.png", refdisp)
fitdisp = oedepict.OE2DMolDisplay(fitmol2D, opts)
oedepict.OERenderMolecule("OEPrepareAlignedDepictionFrom3D-fit-2D.png", fitdisp)
|
|
|
3D reference molecule |
3D molecule overlay |
3D fit molecule |
|
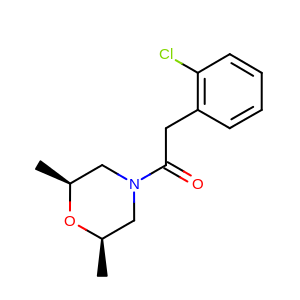
|
|
2D reference molecule (generated by OEPrepareDepictionFrom3D) |
2D fit molecule (generated by OEPrepareAlignedDepictionFrom3D) |
bool OEPrepareAlignedDepictionFrom3D(OEChem::OEMolBase& fitmol2D,
const OEChem::OEMolBase& fitmol3D,
const OEShapeQueryDisplay& qdisp);
Generates the 2D coordinates of a molecule driven by the 3D overlay of the shape query display object.
- fitmol2D
The molecule for which 2D coordinates are generated.
- refmol2D
The 2D molecule whose orientation drives the 2D coordinate generation.
- qdisp
The OEShapeQueryDisplay object that drives the layout of the 2D coordinate generation.
See also
OEShapeQueryDisplay class