Depicting Active Site Interactions
A program that depicts receptor-ligand interactions.
See also
Visualizing Protein-Ligand Interactions OpenEye Python Cookbook example
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> python complex2img.py --help
will generate the following output:
[-complex] <input> [-out] <output image>
Simple parameter list
-height : Height of output image
-width : Width of output image
SplitMolComplex options :
-ligandname : Ligand name
input/output options
-complex : Input filename of the protein complex
-out : Output filename
molecule display options
-aromstyle : Aromatic ring display style
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Depicts the interactions of an active site
#############################################################################
import sys
from openeye import oechem
from openeye import oedepict
from openeye import oegrapheme
def main(argv=[__name__]):
itf = oechem.OEInterface()
oechem.OEConfigure(itf, InterfaceData)
oedepict.OEConfigureImageWidth(itf, 900.0)
oedepict.OEConfigureImageHeight(itf, 600.0)
oedepict.OEConfigure2DMolDisplayOptions(itf, oedepict.OE2DMolDisplaySetup_AromaticStyle)
oechem.OEConfigureSplitMolComplexOptions(itf, oechem.OESplitMolComplexSetup_LigName)
if not oechem.OEParseCommandLine(itf, argv):
return 1
iname = itf.GetString("-complex")
oname = itf.GetString("-out")
ifs = oechem.oemolistream()
if not ifs.open(iname):
oechem.OEThrow.Fatal("Cannot open input file!")
ext = oechem.OEGetFileExtension(oname)
if not oedepict.OEIsRegisteredImageFile(ext):
oechem.OEThrow.Fatal("Unknown image type!")
ofs = oechem.oeofstream()
if not ofs.open(oname):
oechem.OEThrow.Fatal("Cannot open output file!")
complexmol = oechem.OEGraphMol()
if not oechem.OEReadMolecule(ifs, complexmol):
oechem.OEThrow.Fatal("Unable to read molecule from %s" % iname)
if not oechem.OEHasResidues(complexmol):
oechem.OEPerceiveResidues(complexmol, oechem.OEPreserveResInfo_All)
# Separate ligand and protein
sopts = oechem.OESplitMolComplexOptions()
oechem.OESetupSplitMolComplexOptions(sopts, itf)
ligand = oechem.OEGraphMol()
protein = oechem.OEGraphMol()
water = oechem.OEGraphMol()
other = oechem.OEGraphMol()
pfilter = sopts.GetProteinFilter()
wfilter = sopts.GetWaterFilter()
sopts.SetProteinFilter(oechem.OEOrRoleSet(pfilter, wfilter))
sopts.SetWaterFilter(oechem.OEMolComplexFilterFactory(
oechem.OEMolComplexFilterCategory_Nothing))
oechem.OESplitMolComplex(ligand, protein, water, other, complexmol, sopts)
if ligand.NumAtoms() == 0:
oechem.OEThrow.Fatal("Cannot separate complex!")
# Perceive interactions
asite = oechem.OEInteractionHintContainer(protein, ligand)
if not asite.IsValid():
oechem.OEThrow.Fatal("Cannot initialize active site!")
asite.SetTitle(ligand.GetTitle())
oechem.OEPerceiveInteractionHints(asite)
oegrapheme.OEPrepareActiveSiteDepiction(asite)
# Depict active site with interactions
width, height = oedepict.OEGetImageWidth(itf), oedepict.OEGetImageHeight(itf)
image = oedepict.OEImage(width, height)
cframe = oedepict.OEImageFrame(image, width * 0.80, height, oedepict.OE2DPoint(0.0, 0.0))
lframe = oedepict.OEImageFrame(image, width * 0.20, height,
oedepict.OE2DPoint(width * 0.80, 0.0))
opts = oegrapheme.OE2DActiveSiteDisplayOptions(cframe.GetWidth(), cframe.GetHeight())
oedepict.OESetup2DMolDisplayOptions(opts, itf)
adisp = oegrapheme.OE2DActiveSiteDisplay(asite, opts)
oegrapheme.OERenderActiveSite(cframe, adisp)
lopts = oegrapheme.OE2DActiveSiteLegendDisplayOptions(10, 1)
oegrapheme.OEDrawActiveSiteLegend(lframe, adisp, lopts)
oedepict.OEWriteImage(oname, image)
return 0
#############################################################################
# INTERFACE
#############################################################################
InterfaceData = '''
!BRIEF [-complex] <input> [-out] <output image>
!CATEGORY "input/output options :"
!PARAMETER -complex
!ALIAS -c
!TYPE string
!KEYLESS 1
!REQUIRED true
!VISIBILITY simple
!BRIEF Input filename of the protein complex
!END
!PARAMETER -out
!ALIAS -o
!TYPE string
!REQUIRED true
!KEYLESS 2
!VISIBILITY simple
!BRIEF Output filename
!END
!END
'''
if __name__ == "__main__":
sys.exit(main(sys.argv))
See also
OESplitMolComplexfunction in OEChem TKOEInteractionHintContainer class in OEChem TK
OEPerceiveInteractionHintsfunction in OEChem TKOEPrepareActiveSiteDepictionfunctionOE2DActiveSiteDisplay class and
OERenderActiveSitefunctionOEDrawActiveSiteLegendfunction
Examples
prompt> python complex2img.py -complex pdb1br6.ent -out 1br6-PT1.png
will generate the image shown in Figure: Example of depicting the receptor-ligand interaction (PDB: 1br6).
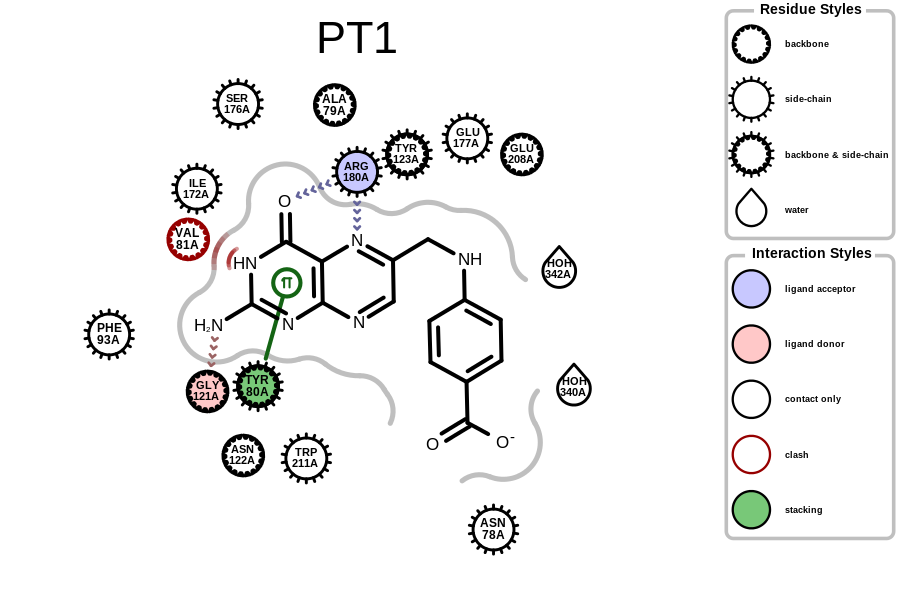
Example of depicting the receptor-ligand interactions (PDB: 1br6)
prompt> python complex2img.py -complex pdb1eoc.ent -out 1eoc-4NC.png
will generate the image shown in Figure: Example of depicting the receptor-ligand interaction (PDB: 1eoc).
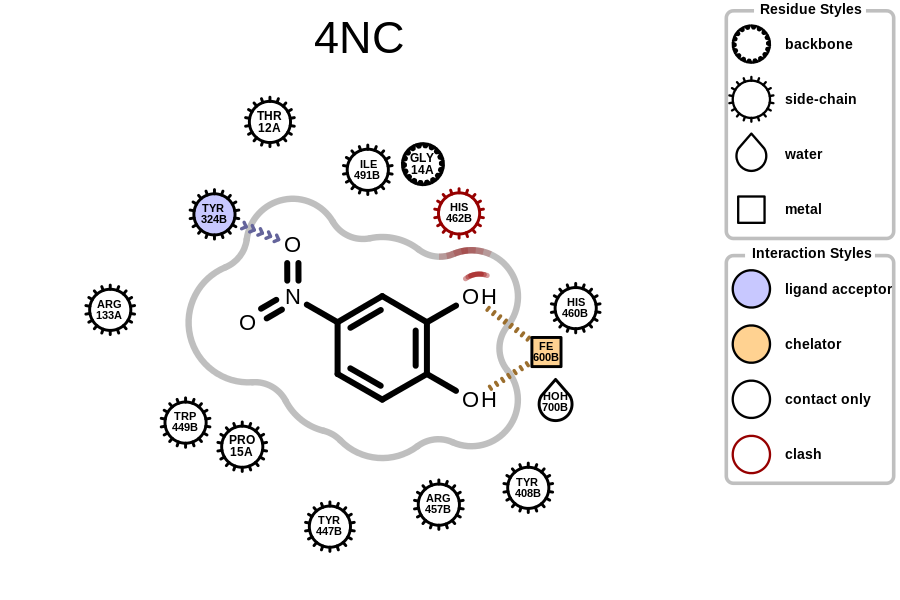
Example of depicting the receptor-ligand interactions (PDB: 1eoc)