Depicting Shape and Color Atom Overlaps
A program that depicts the shape and the color overlap between a 3D reference structure and a sets of pre-aligned 3D fit molecules. It is implemented to visualize the output of ROCS but can be used to visualize the shape and color similarities of any molecules that have been pre-aligned with other applications.
Note
OEShape TK license is not required to run this example.
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> python shapeoverlap2pdf.py --help
will generate the following output:
[-in] <input> [-out] <output pdf>
Simple parameter list
input/output options:
-in : Input molecule filename
-out : Output image filename
general options:
-maxhits : Maximum number of hits depicted
Code
Download code
#!/usr/bin/env python
# (C) 2022 Cadence Design Systems, Inc. (Cadence)
# All rights reserved.
# TERMS FOR USE OF SAMPLE CODE The software below ("Sample Code") is
# provided to current licensees or subscribers of Cadence products or
# SaaS offerings (each a "Customer").
# Customer is hereby permitted to use, copy, and modify the Sample Code,
# subject to these terms. Cadence claims no rights to Customer's
# modifications. Modification of Sample Code is at Customer's sole and
# exclusive risk. Sample Code may require Customer to have a then
# current license or subscription to the applicable Cadence offering.
# THE SAMPLE CODE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
# EXPRESS OR IMPLIED. OPENEYE DISCLAIMS ALL WARRANTIES, INCLUDING, BUT
# NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
# PARTICULAR PURPOSE AND NONINFRINGEMENT. In no event shall Cadence be
# liable for any damages or liability in connection with the Sample Code
# or its use.
#############################################################################
# Depicts shape and color overlap between a 3D reference structure and
# a sets of 3D fit molecules. The molecules have to be pre-aligned.
# The first molecule is expected be the reference.
#############################################################################
import sys
from openeye import oechem
from openeye import oedepict
from openeye import oegrapheme
from openeye import oeshape
def main(argv=[__name__]):
itf = oechem.OEInterface(InterfaceData)
if not oechem.OEParseCommandLine(itf, argv):
return 1
iname = itf.GetString("-in")
oname = itf.GetString("-out")
maxhits = itf.GetInt("-maxhits")
ext = oechem.OEGetFileExtension(oname)
if not oedepict.OEIsRegisteredMultiPageImageFile(ext):
oechem.OEThrow.Fatal("Unknown multipage image type!")
ifs = oechem.oemolistream()
if not ifs.open(iname):
oechem.OEThrow.Fatal("Cannot open input molecule file!")
refmol = oechem.OEGraphMol()
if not oechem.OEReadMolecule(ifs, refmol):
oechem.OEThrow.Fatal("Cannot read reference molecule!")
ropts = oedepict.OEReportOptions(3, 1)
ropts.SetHeaderHeight(40.0)
ropts.SetFooterHeight(20.0)
report = oedepict.OEReport(ropts)
cff = oeshape.OEColorForceField()
cff.Init(oeshape.OEColorFFType_ImplicitMillsDean)
cffdisplay = oegrapheme.OEColorForceFieldDisplay(cff)
qopts = GetShapeQueryDisplayOptions()
sopts = GetShapeOverlapDisplayOptions()
copts = GetColorOverlapDisplayOptions()
refdisp = oegrapheme.OEShapeQueryDisplay(refmol, cff, qopts)
dots = oechem.OEDots(100, 10, "shape overlaps")
for fitmol in ifs.GetOEGraphMols():
if maxhits > 0 and dots.GetCounts() >= maxhits:
break
dots.Update()
maincell = report.NewCell()
grid = oedepict.OEImageGrid(maincell, 1, 3)
grid.SetMargins(5.0)
grid.SetCellGap(5.0)
# TITLE + SCORE GRAPH + QUERY
cell = grid.GetCell(1, 1)
cellw, cellh = cell.GetWidth(), cell.GetHeight()
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Bold, 10,
oedepict.OEAlignment_Left, oechem.OEBlack)
pos = oedepict.OE2DPoint(10.0, 10.0)
cell.DrawText(pos, fitmol.GetTitle(), font, cell.GetWidth())
rframe = oedepict.OEImageFrame(cell, cellw, cellh * 0.35,
oedepict.OE2DPoint(0.0, cellh * 0.10))
mframe = oedepict.OEImageFrame(cell, cellw, cellh * 0.50,
oedepict.OE2DPoint(0.0, cellh * 0.50))
RenderScoreRadial(rframe, fitmol)
oegrapheme.OERenderShapeQuery(mframe, refdisp)
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Bold, 8,
oedepict.OEAlignment_Center, oechem.OEGrey)
pos = oedepict.OE2DPoint(20.0, 10.0)
mframe.DrawText(pos, "query", font)
oedepict.OEDrawCurvedBorder(mframe, oedepict.OELightGreyPen, 10.0)
odisp = oegrapheme.OEShapeOverlapDisplay(refdisp, fitmol, sopts, copts)
# SHAPE OVERLAP
cell = grid.GetCell(1, 2)
oegrapheme.OERenderShapeOverlap(cell, odisp)
RenderScore(cell, fitmol, "ROCS_ShapeTanimoto", "Shape Tanimoto")
# COLOR OVERLAP
cell = grid.GetCell(1, 3)
oegrapheme.OERenderColorOverlap(cell, odisp)
RenderScore(cell, fitmol, "ROCS_ColorTanimoto", "Color Tanimoto")
oedepict.OEDrawCurvedBorder(maincell, oedepict.OELightGreyPen, 10.0)
dots.Total()
cffopts = oegrapheme.OEColorForceFieldLegendDisplayOptions(1, 6)
for header in report.GetHeaders():
oegrapheme.OEDrawColorForceFieldLegend(header, cffdisplay, cffopts)
oedepict.OEDrawCurvedBorder(header, oedepict.OELightGreyPen, 10.0)
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Default, 12,
oedepict.OEAlignment_Center, oechem.OEBlack)
for idx, footer in enumerate(report.GetFooters()):
oedepict.OEDrawTextToCenter(footer, "-" + str(idx + 1) + "-", font)
oedepict.OEWriteReport(oname, report)
return 0
def AddCommonDisplayOptions(opts):
opts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
opts.SetAtomLabelFontScale(1.5)
pen = oedepict.OEPen(oechem.OEBlack, oechem.OEBlack, oedepict.OEFill_Off, 1.5)
opts.SetDefaultBondPen(pen)
def GetShapeQueryDisplayOptions():
qopts = oegrapheme.OEShapeQueryDisplayOptions()
AddCommonDisplayOptions(qopts)
arcpen = oedepict.OEPen(oedepict.OELightGreyPen)
qopts.SetSurfaceArcFxn(oegrapheme.OEDefaultArcFxn(arcpen))
qopts.SetDepictOrientation(oedepict.OEDepictOrientation_Square)
return qopts
def GetShapeOverlapDisplayOptions():
sopts = oegrapheme.OEShapeOverlapDisplayOptions()
AddCommonDisplayOptions(sopts)
arcpen = oedepict.OEPen(oechem.OEGrey, oechem.OEGrey, oedepict.OEFill_Off, 1.0, 0x1111)
sopts.SetQuerySurfaceArcFxn(oegrapheme.OEDefaultArcFxn(arcpen))
sopts.SetOverlapColor(oechem.OEColor(110, 110, 190))
return sopts
def GetColorOverlapDisplayOptions():
copts = oegrapheme.OEColorOverlapDisplayOptions()
AddCommonDisplayOptions(copts)
arcpen = oedepict.OEPen(oechem.OEGrey, oechem.OEGrey, oedepict.OEFill_Off, 1.0, 0x1111)
copts.SetQuerySurfaceArcFxn(oegrapheme.OEDefaultArcFxn(arcpen))
return copts
def GetScore(mol, sdtag):
if oechem.OEHasSDData(mol, sdtag):
return float(oechem.OEGetSDData(mol, sdtag))
return 0.0
def RenderScoreRadial(image, mol):
sscore = max(min(GetScore(mol, "ROCS_ShapeTanimoto"), 1.0), 0.00)
cscore = max(min(GetScore(mol, "ROCS_ColorTanimoto"), 1.0), 0.00)
if sscore > 0.0 or cscore > 0.0:
scores = oechem.OEDoubleVector([sscore, cscore])
oegrapheme.OEDrawROCSScores(image, scores)
def RenderScore(image, mol, sdtag, label):
score = GetScore(mol, sdtag)
if score == 0.0:
return
w, h = image.GetWidth(), image.GetHeight()
frame = oedepict.OEImageFrame(image, w, h * 0.10, oedepict.OE2DPoint(0.0, h * 0.90))
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Default, 9,
oedepict.OEAlignment_Center, oechem.OEBlack)
oedepict.OEDrawTextToCenter(frame, label + " = " + str(score), font)
#############################################################################
# INTERFACE
#############################################################################
InterfaceData = '''
!BRIEF [-in] <input> [-out] <output pdf>
!CATEGORY "input/output options:" 0
!PARAMETER -in
!ALIAS -i
!TYPE string
!REQUIRED true
!KEYLESS 1
!VISIBILITY simple
!BRIEF Input molecule filename
!DETAIL
The first molecule in the file is expected to be the reference
molecule
!END
!PARAMETER -out
!ALIAS -o
!TYPE string
!REQUIRED true
!KEYLESS 2
!VISIBILITY simple
!BRIEF Output image filename
!END
!END
!CATEGORY "general options:" 1
!PARAMETER -maxhits
!ALIAS -mhits
!TYPE int
!REQUIRED false
!DEFAULT 0
!LEGAL_RANGE 0 500
!VISIBILITY simple
!BRIEF Maximum number of hits depicted
!DETAIL
The default of 0 means there is no limit.
!END
!END
'''
if __name__ == "__main__":
sys.exit(main(sys.argv))
See also
OEColorForceField class in the OEShape TK
OEShapeQueryDisplay and OEShapeQueryDisplayOptions classes
OERenderShapeQueryfunctionOEShapeOverlapDisplay, OEShapeOverlapDisplayOptions, and OEColorOverlapDisplayOptions classesOERenderShapeOverlapfunctionOERenderColorOverlapfunctionOEDrawColorForceFieldLegendfunctionOEDrawROCSScoresfunction
Examples
prompt> python shapeoverlap2pdf.py -in rocs.sdf -out overlaps.pdf
page 1 |
page 2 |
page 3 |
|---|---|---|
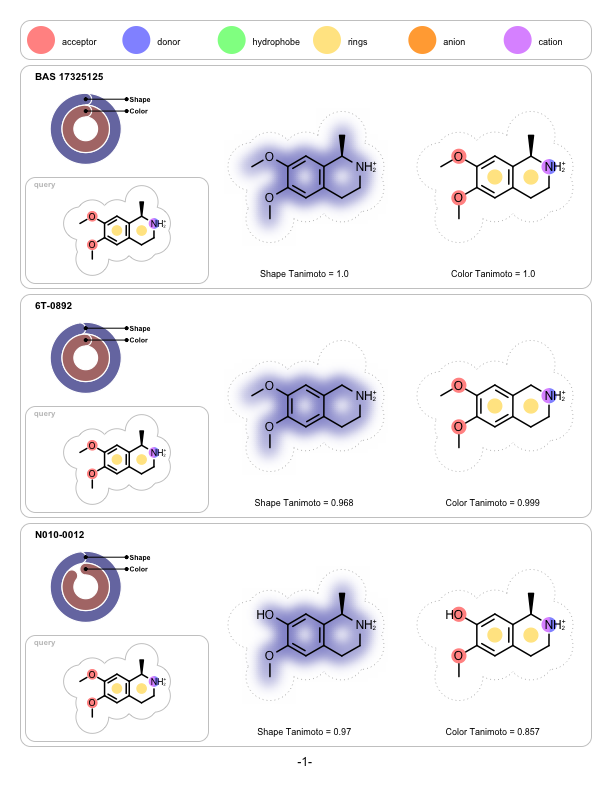
|
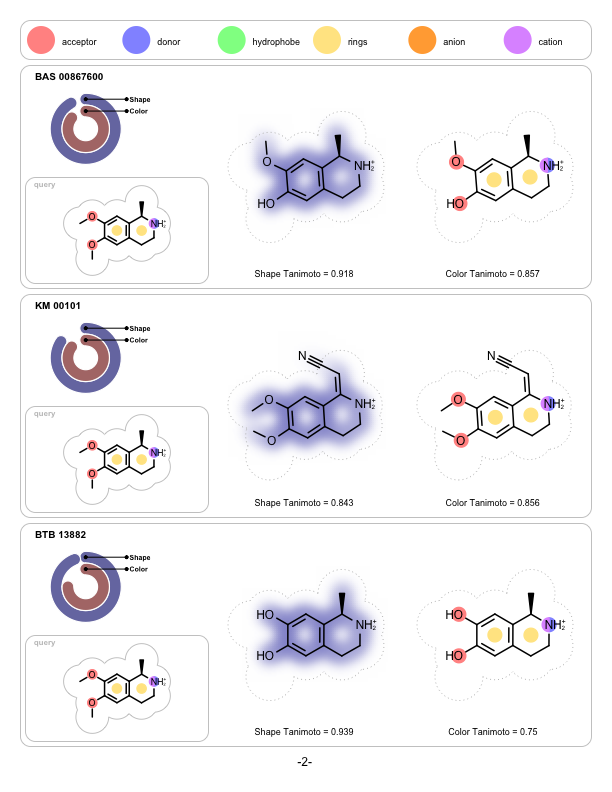
|
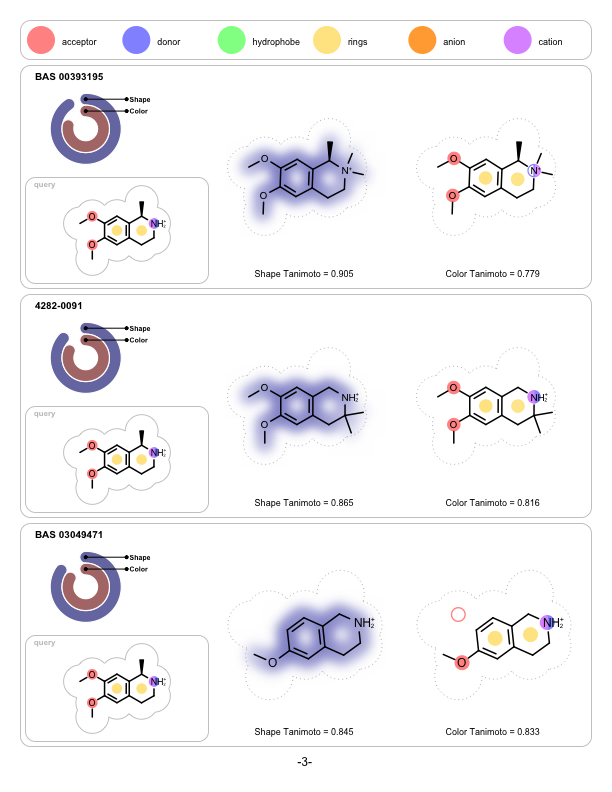
|