Overview
vBROOD is the graphical interface that ships with BROOD. It gives access to all the command line features as well as some GUI-only features, such as query editing, and placing constraint and color atoms.
Upon completion, BROOD clusters the resultant molecules. vBROOD presents the cluster heads for viewing; the full clusters can be explored in VIDA (see Viewing Results in VIDA).
Most BROOD options can be changed from within vBROOD. vBROOD can easily run BROOD in multiprocessor mode; however, it cannot run BROOD across a cluster.
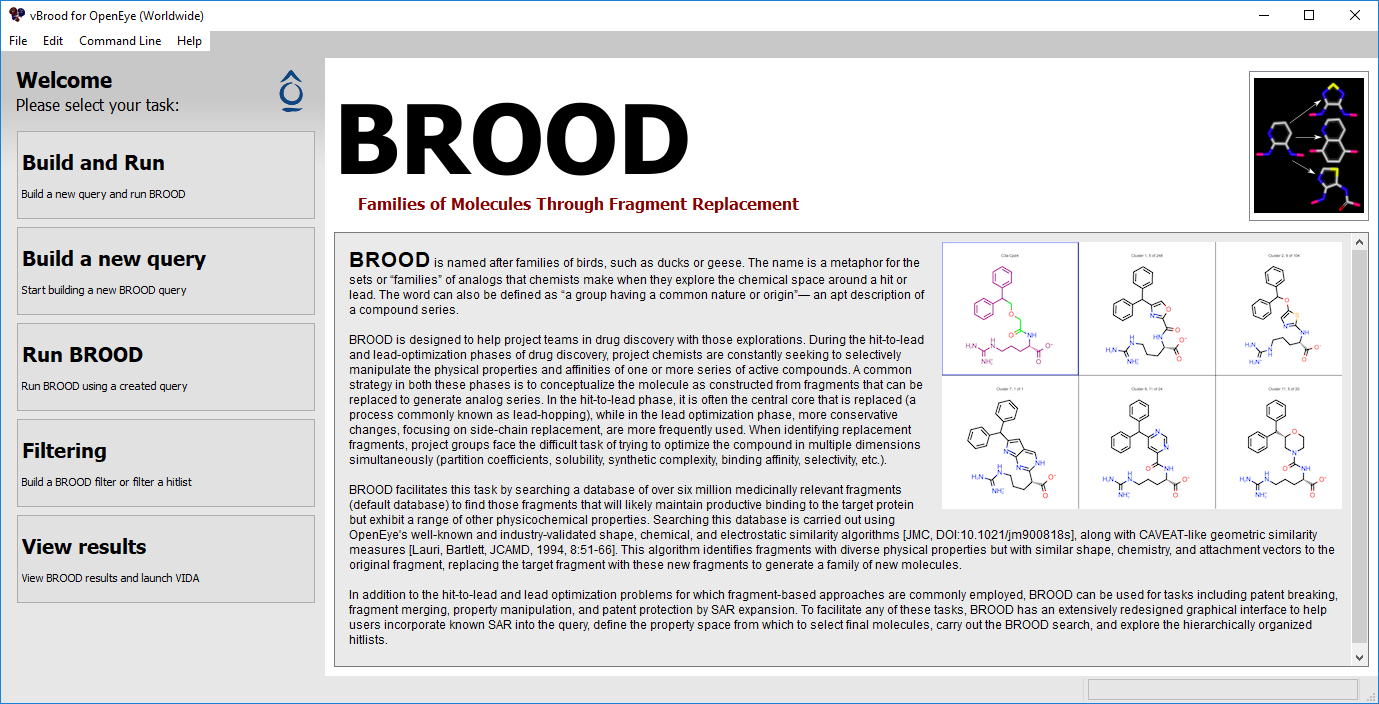
Figure 1. The vBROOD task screen.
vBROOD offers five tasks to help run BROOD:
Build and Run: Creates a query and immediately starts a BROOD run.
Build a New Query: Creates and edits a query to save and send to a BROOD run.
Run BROOD: Runs BROOD with a query.
View Results: Browses results and launches the BROOD interface in VIDA.