Analyzing SZMAP Results
Determining the Ligand Displacement Region
The results can be processed with GridComp to break-out the region of the apo grid that has been displaced by the ligand.
> grid_comp -op lig_disp -i 4std_stbl.oeb.gz -o 4std_stbl_disp.oeb.gz
While not required, these displacement grids can be useful. For example, they can be used to identify precisely which waters are displaced by each of a series of substituents. They also make it easy to determine the volume of water displaced by the ligand.
Analyzing Grid Results
SZMAP grid results can be visualized in VIDA using the WaterColor VIDA Extension (sets grid styles for viewing—the link leads to information about the extension) and the Water Orientation VIDA Extension (shows water orientational preferences). If VIDA is in your command search path, the following command will open VIDA and load the file 4std_stbl_disp.oeb.gz. VIDA can also be launched by clicking on the VIDA icon and the file can then be loaded using the Open… command in the File menu. You can also drag and drop molecule files onto a VIDA instance.
> vida 4std_stbl_disp.oeb.gz
Note
VIDA extensions need to be installed before they can be used. For instructions, see Installing VIDA Extensions.
The neutral difference (water - uncharged) grids map the polarity throughout the site and the stabilization grids describe the binding reaction (complex - apo - ligand): they have negative energies where water enhances binding affinity and positive where it decreases binding affinity.
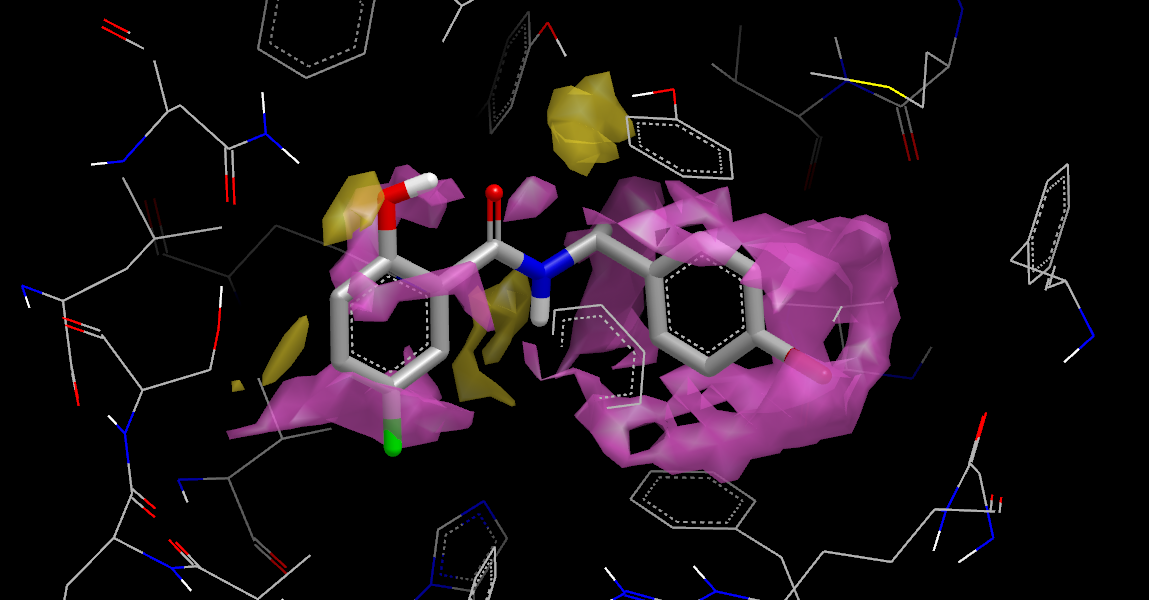
Displaced Neutral Difference Free Energy
The figure Displaced Neutral Difference Free Energy was generated by selecting 4std_stbl_disp.oeb.gz in VIDA’s list window and running the WaterColor VIDA Extension to setup the contour levels and display styles, then opening the lig_disp:apo:PROTEIN… item in the list window and displaying the neut_diff_apo_free_energy_grid. You must run WaterColor on each new SZMAP results file opened in VIDA. See chapter WaterColor VIDA Extension for more detailed instructions.
The figure Displaced Neutral Difference Free Energy shows that the ligand displaces regions with both negative (yellow) and positive (purple) neutral difference free energies. These energies represent a polarity scale: negative regions favor polar groups while positive regions favor non-polar groups. Note that the hydroxyl oxygen nicely substitutes for solvent that acts as an H-bond acceptor. The region in the upper center, on the other hand, contains solvent interacting with a pair of tyrosine hydroxyls that is displaced but not replaced with a similar group. This suggests placing a polar group into this site.
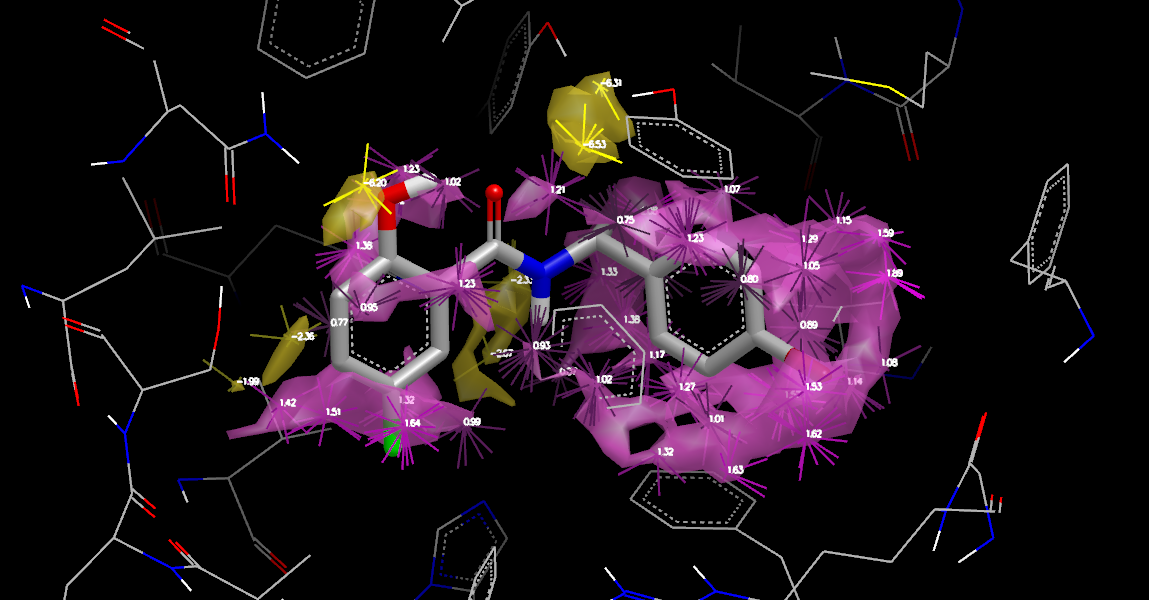
Orientations and Displaced Neutral Difference Free Energy
The Water Orientation VIDA Extension will display the most probable water orientations for each point in the calculation, filtered to show only the dominant points (local minima and maxima). It makes it easy to determine where waters donate or accept H-bonds and where waters do not make any significant interactions. Open the control panel by selecting the Extensions >> Water Orientation menu in VIDA (it will remain open until you close it).
The figure
Orientations and Displaced Neutral Difference Free Energy
adds information indicating both the magnitude of the free energies
and the geometry that must be taken into account in positioning a
substituent. To generate the figure, set the Apply to: popup
in the Water Orientation VIDA Extension
to 4std_stbl_disp.oeb.gz/apo(lig_disp)—the ligand displacement apo
results—and press the Update button.
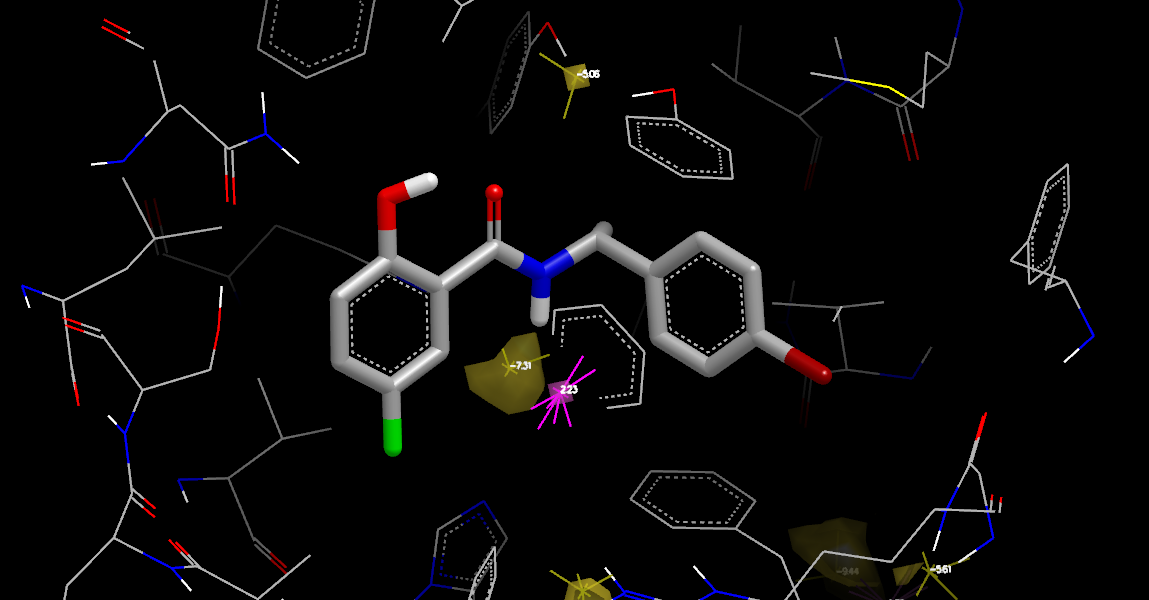
Complex Neutral Difference Free Energy
The next stage is to examine the protein-ligand complex grid, illustrated in
Complex Neutral Difference Free Energy.
First, undisplay all grids from the lig_disp:apo:PROTEIN… entry
in VIDA’s list window,
along with the 4std_stbl_disp.oeb.gz/apo(lig_disp) entry from
the previous orientation display.
Next, open the complex:PROTEIN… section of VIDAs list window
to reveal the set of grids calculated for the complex.
Select and display the neut_diff_free_energy_grid.
Finally, set the Water Orientation VIDA Extension to applied to 4std_stbl_disp.oeb.gz/complex
and press Update.
The display shows two regions of negative free energy: a region at the bottom were
solvent interacts with two histidines and the amide NH of the ligand,
and a smaller region at the top, just beyond the displaced region, that
interacts with the previously mentioned tyrosines, along with one region of
positive free energy.
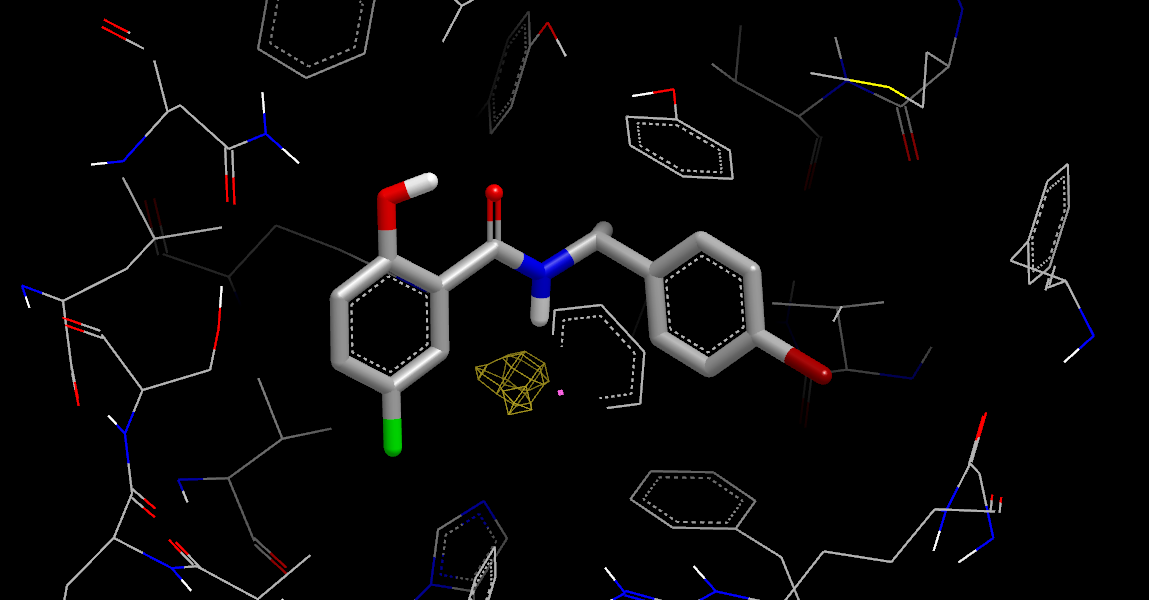
Stabilization Neutral Difference Free Energy
Looking at the Stabilization Neutral Difference Free Energy (by undisplaying any other grids or orientations and then opening the stabilization:PROTEIN… item in the VIDA list window and displaying the neut_diff_stbl_free_energy_grid), it is clear that only the lower region is stabilizing the bound ligand, adding weight to the idea of displacing water in the upper region.