Viewing Results in VIDA
VIDA is OpenEye’s molecular visualization tool. Although vBROOD provides some facility for browsing results, VIDA allows for more full-featured cluster browsing. vBROOD can launch the Results Browser tool from either the last page of a BROOD run or from the Results Browser in vBROOD. Click “Open in VIDA” to launch VIDA with the BROOD Cluster Browser loaded.
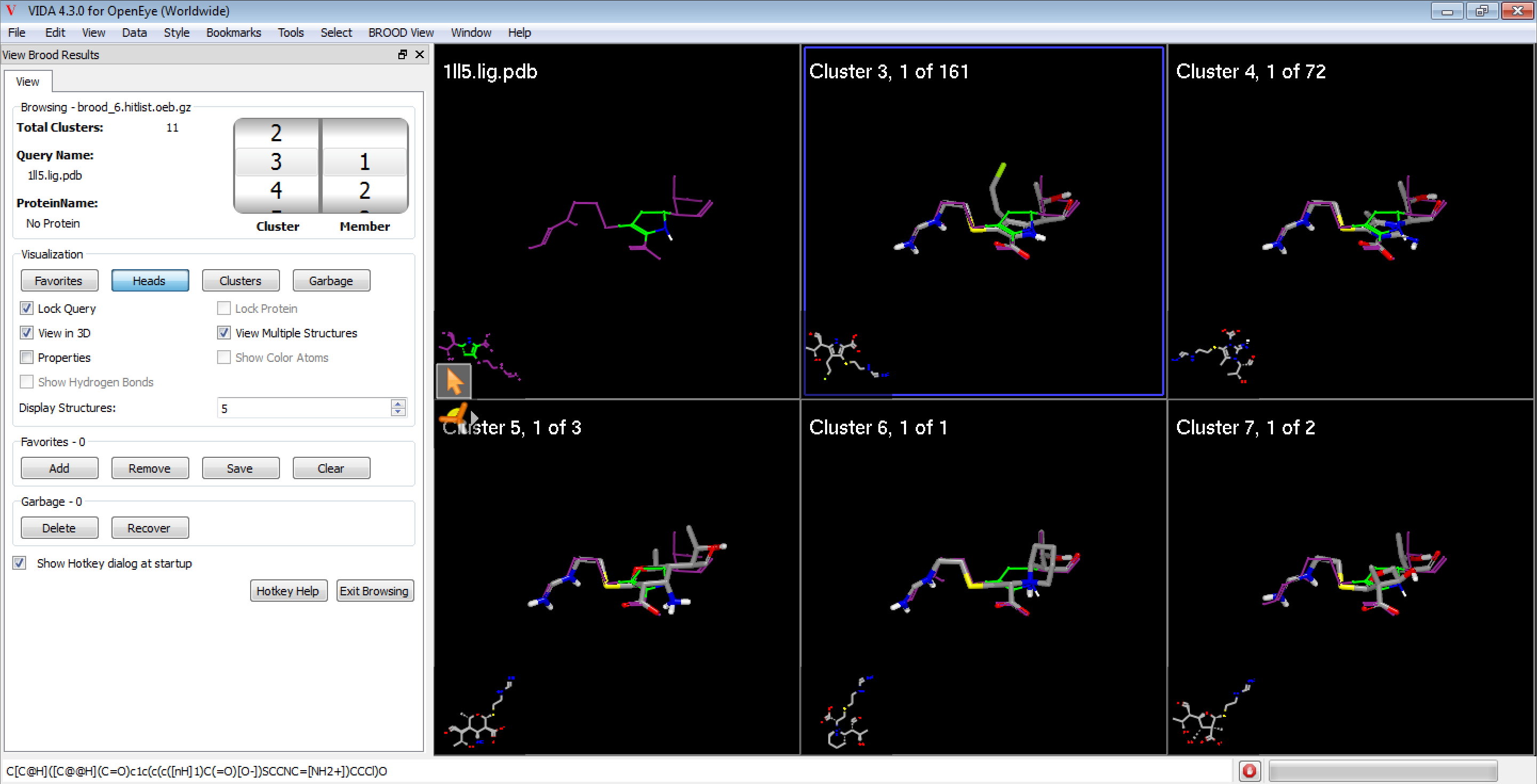
Figure 1. The BROOD Cluster Browser in VIDA.
Getting Started
To open the BROOD Cluster Browser in VIDA, click “Open in VIDA” from either the Browse Results wizard or from the Run BROOD page in the either the Run BROOD wizard or the Build and Run wizard. If VIDA cannot be found or has not been previously launched, vBROOD will prompt you to locate it. The path to VIDA can also be specified in the vBROOD preferences dialog.
When vBROOD launches VIDA with the cluster browser, it first displays the new user interface and then loads the BROOD results. After the results have been loaded, the cluster browser will open the Hotkey Help dialog box displaying the various shortcut keys that can be used in the 3D window. This dialog can be closed by clicking “OK” and reopened by clicking on the Hotkey Help button in the BROOD Cluster Browser window.
The BROOD Cluster Browser window displays information about the hit list as well as tools for controlling the visualization of results, marking favorite molecules, and hiding other molecules in the Trash.
The BROOD Cluster Browser also adds a new menu, BROOD Viewer, which allows for loading and reloading BROOD results.
Viewing Clusters
The BROOD Viewer has four sections: Browsing Information, Visualization Control, Favorites, and Garbage.
Browsing
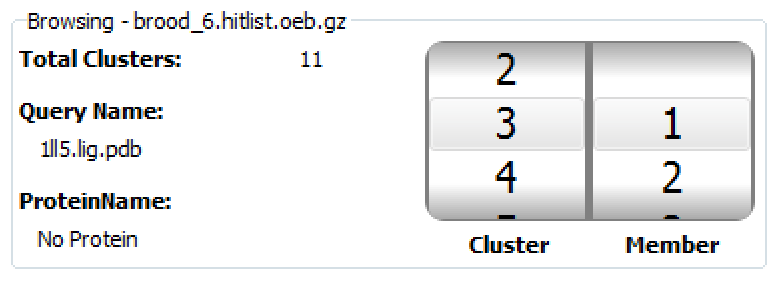
Figure 2. The Browsing section of the BROOD Viewer.
The Browsing Information section provides general information about the displayed hit list. It shows which hit list, query, and protein are being displayed, information about the clusters in the file, and the current cluster being viewed.
The browsing window also contains a spinner that displays which cluster and which member you are currently browsing. The left spinner lists the clusters. If the selected cluster is blue, you are navigating the cluster heads. The right spinner shows the members of the current cluster. If the selected number in the right spinner is highlighted in blue, you are navigating with the cluster. This spinner can be useful for keyboard navigation.
Visualization
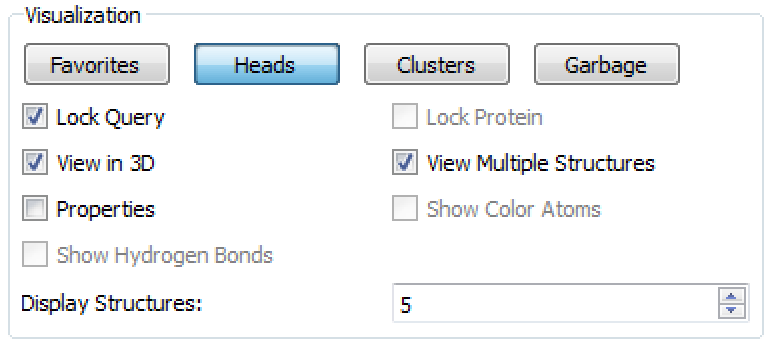
Figure 3. The Visualization section of the BROOD Viewer.
The Visualization section contains the primary set of controls. The four buttons—Favorites, Heads, Clusters and Garbage—control which structures are being displayed in the main view.
Heads navigates among the cluster heads.
Clusters switches to browsing the current cluster members.
Favorites displays any structures marked as favorites by the user.
Garbage displays any molecules hidden in the garbage through the use of the Delete button.
The Visualization section also provides checkboxes to control the view.
Lock Query displays the query alongside a result in each pane in 3D.
Lock Protein displays the protein alongside a result in each pane in 3D.
View in 3D toggles the cluster display between 3D and 2D.
View Multiple Structures toggles between viewing a single result structure and a grid of structures.
Properties toggles the display of a context-sensitive spreadsheet which displays medicinal chemistry properties for the current cluster.
Show Color Atoms turns on the display of color atoms, if enabled by Attach Color Atoms under Advanced Options in vBROOD.
Show Hydrogen Bonds toggles the display of hydrogen bonds between the protein and the result molecules.
Display Structures controls the number of structures displayed in Multiple Structure view.
Favorites and Garbage
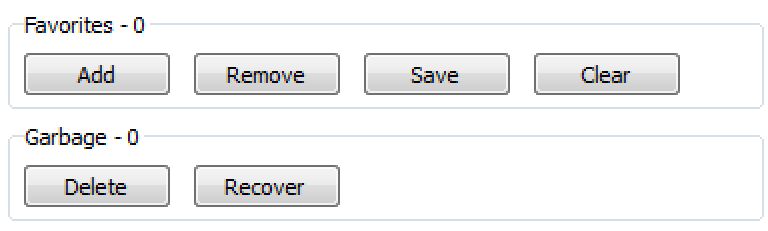
Figure 4. The Favorites and Garbage sections.
The Favorites section allows users to tag individual molecules as Favorites for further follow-up or saving. The number of favorites are displayed in the caption. Below the caption are four buttons.
Add puts the current structure (surrounded by the blue box) into the Favorites list.
Remove takes the current structure (surrounded by the blue box) out of the Favorites list.
Save gives a prompt for a file name, then saves the Favorites list to disk.
Clear empties the Favorites list, moving all molecules back to their original location.
The Garbage section provides a way to hide structures from view. They are still available but are hidden from normal browsing. The number of structures in Garbage is displayed in the heading. The Garbage can be browsed by clicking View Garbage. Clicking Recover while viewing a structure in the Garbage will put the molecule back into its original cluster.
Keyboard Control
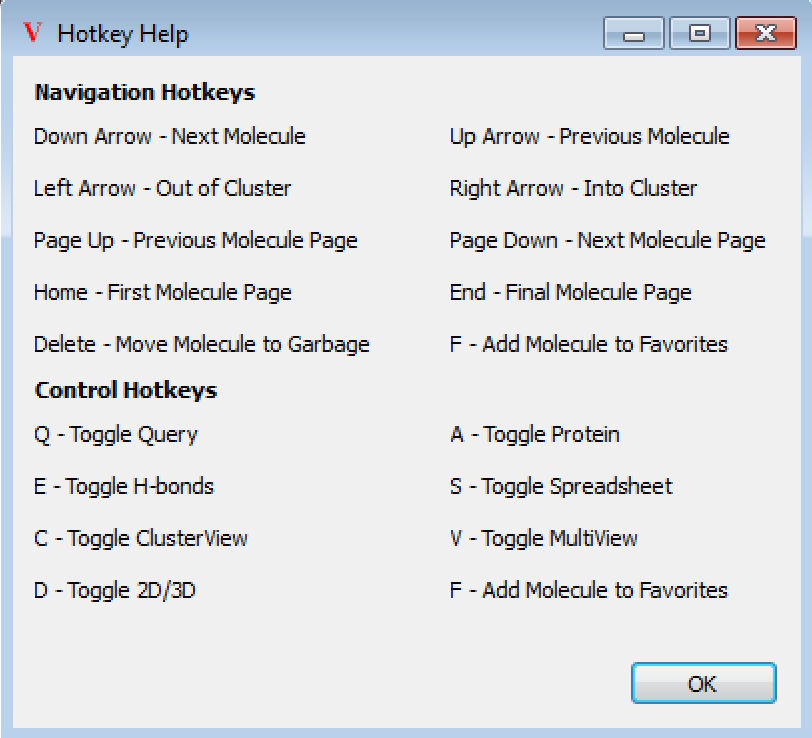
Figure 5. The Hotkey Help dialog box.
The VIDA Cluster Browser script has key bindings for many common operations. The keyboard operations fall into two categories: cluster navigation and display control.
Display Control Hotkeys
Many of the controls available in the Visualization section are also keyboard accessible.
Q toggles display of the query.
E toggles the hydrogen bond display.
C toggles between navigation of the heads and the clusters.
D toggles between the 2D and 3D display.
A toggles display of the protein (if any).
S toggles display of the spreadsheet.
V toggles display of multiple structures.
F adds a molecule to favorites.