Overview
sitehopper_search is used to search a database of OEDesignUnits similar to a query OEDesignUnit.
Note
To use GPU prescreen, an NVIDIA GPU is required to run sitehopper_search. See GPU Prerequisites for details. If not GPU is available, search will revert to CPU only search.
Basic Search Process
Search consists of several phases.
GPU prescreen: (assuming a compatible GPU is available and
-use_gpuis true). Data is loaded into memory and then searched, keeping the top 5000 hits to send on to CPU Search.CPU Search: Use
-ncpuCPU threads to search. If GPU prescreen, this searches the 5000 results from step 1, otherwise it will search the entire database.Hitlist processing: Hits are extracted from the database, transformed into their final orientation and if
-normalize_and_rescoreis used, that will happen in this step.Writing results files
Output from sitehopper_search
sitehopper_search will create several files:
The .csv file contains scores from the top hits in comma-separated format.
The .oedu file holds the OEDesignUnits for the hits, as well as a surface representation of the binding site. The protein in each OEDesignUnit is tagged with Patch Score, Patch Shape Score, and Patch Color Score and Sequence Similarity. These values can be found in the Proteins spreadsheet window in VIDA.
The .log file contains any warnings or messages from the search, as well as a second copy of the scores.
The .param contains all the parameters used to run search and can be used to re-run the identical search using the
-paramflag.
Scoring in sitehopper_search
sitehopper_search orders hits according Patch Score. Instead of calculating a Color Tanimoto from 0-1, sitehopper_search calculates a color score from 0-3, by multiplying Color Tanimoto by 3. This value is called Patch Color Score. The shape tanimoto is calculated the same as in ShapeTK, and called Patch Shape Score. The resulting Patch Score in sitehopper_search is equal to the sum of the Patch Color and Patch Shape scores, analogous to how the Tanimoto combo score is the sum of Tanimoto shape and color. Patch Score ranges from 0-4, rather than 0-2, with 4.0 being perfect overlap, and 0.0 being no overlap. The shape and color components are output alongside the combo score in the log file, and can also be viewed in the spreadsheet window under the protein tab in VIDA.
Binding site surface visualization
A surface representation of the binding site is included as part of the output design units. An example is shown below:
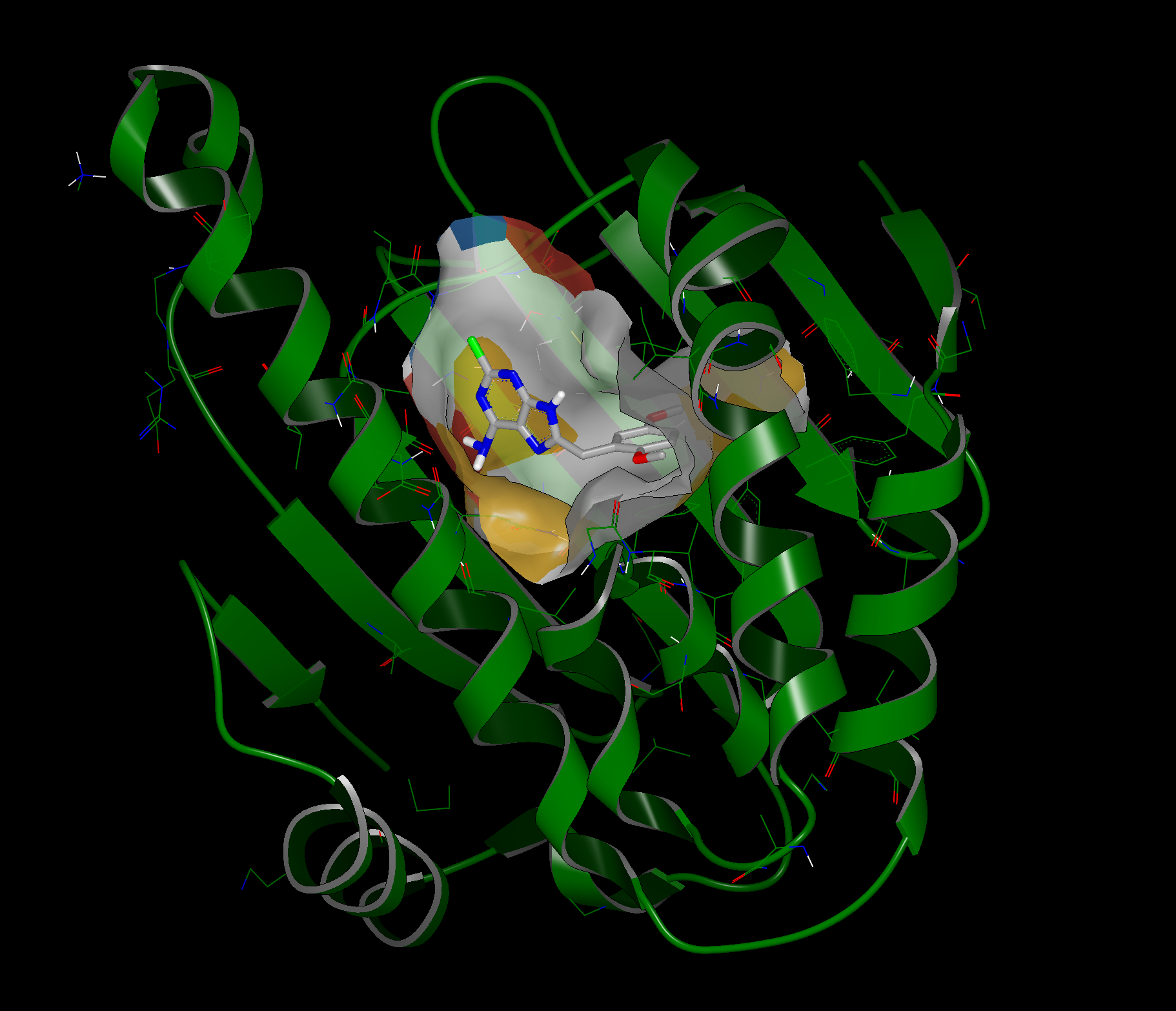
1UYG with binding site surface visualization
The surface represents residues that are close enough to interact with the ligand. The surface is comprised of four colors: white, yellow, red and blue. White corresponds to non-polar residues, yellow to polar residues, red to acidic residues, and blue to basic residues.
Pre-Built SiteHopper Database
Included in the SiteHopper product is a large database which can be searched using sitehopper_search. This database is available for download in a platform-independent format.