CombineReceptors tutorial
Combining receptors is an optional step that can be used to take information found in multiple receptors to create a new, merged, receptor.
combine_receptors is intended to be used in an automated fashion where the output aligned receptors should be included with the original receptors when running POSIT. As such, the output receptor files are automatically named based on the input filenames and only receptors that are capable of merging produce output.
This is particularly helpful when using small bound fragments to try and predict binding poses for larger ligands. Combining receptors works by supplying a list of potential receptors and only receptors that are deemed worthy or merging are output. To be worthy of merging:
It must be possible to align the protein sequences.
The ligands must overlap, but be different enough to indicate that the combined receptor has more information content than each independently.
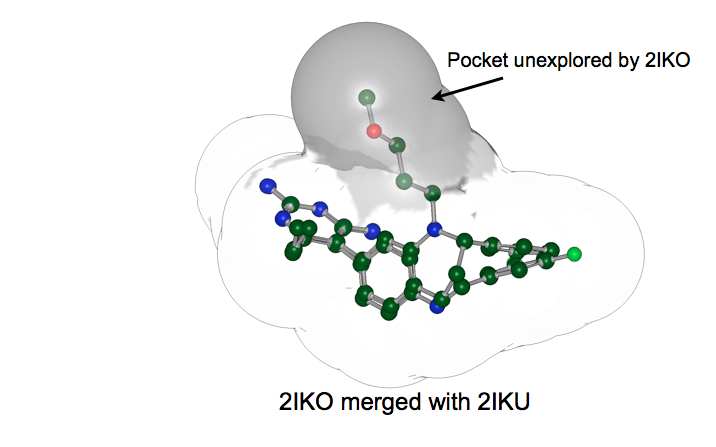
Merged Receptors: Merging the receptors 2IKO and 2IKU capture more potential interaction constraints than either does alone
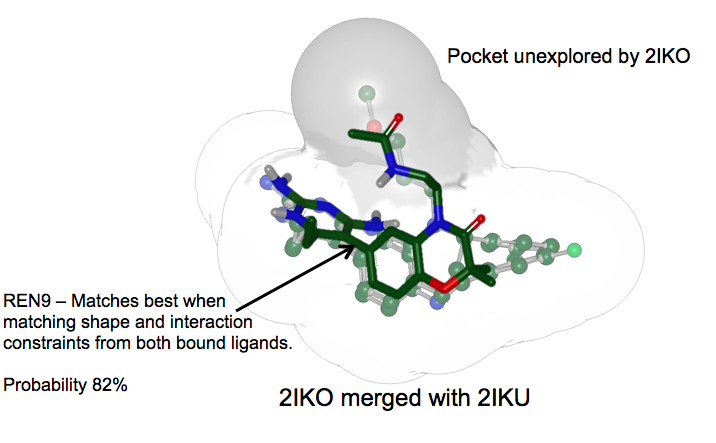
Ligand Posed to Merged Receptors: REN9 is correctly predicted by the merged receptor
For example, consider the merged receptor shown in figure: Merged Receptors. In this case, one of the ligands binds to a pocket that is not present in the other. The expectation is that the combination of both ligands has more information content than either alone.
For each valid merge, two output files are generated, one in each reference frame. See the combine_receptors section for more details.