Release Highlights 2020.2.2
The OEApplications 2020.2.2 is a bug-fix of the OEApplications 2020.1 release, and depends on OEToolkits 2020.2.2. Click on the below links for more information about the bug fixes in this release:
Release Highlights 2020.2
OMEGA: New fragment library
A new enhanced fragment library has been created for use in OMEGA. The new fragment library consists of more than 500,000 fragments that have been generated using the MMFF force field.
This library is designed to reduce the run time of large-scale conformer
generation,
and therefore is not built into OMEGA but is available as a separate download.
To run OMEGA with the enhanced library, simply use the command line flag -addfraglib enhanced_fraglib.oeb.gz.
The fragments have been collated from commonly used electronic databases, such as Enamine Building Blocks. Consequently, dramatic runtime performance improvements can be seen for the Enamine Real dataset, among others, since less time is spent building fragments on the fly. Runtime performance improvements can be seen for all datasets; the Platinum and Enamine Real sets show particular improvement and run ~50% faster (benchmarks run on Ubuntu 16 with Intel(R) Xeon(R) Gold 6128 CPU 3.40GHz and NVIDIA Tesla V100 GPU).
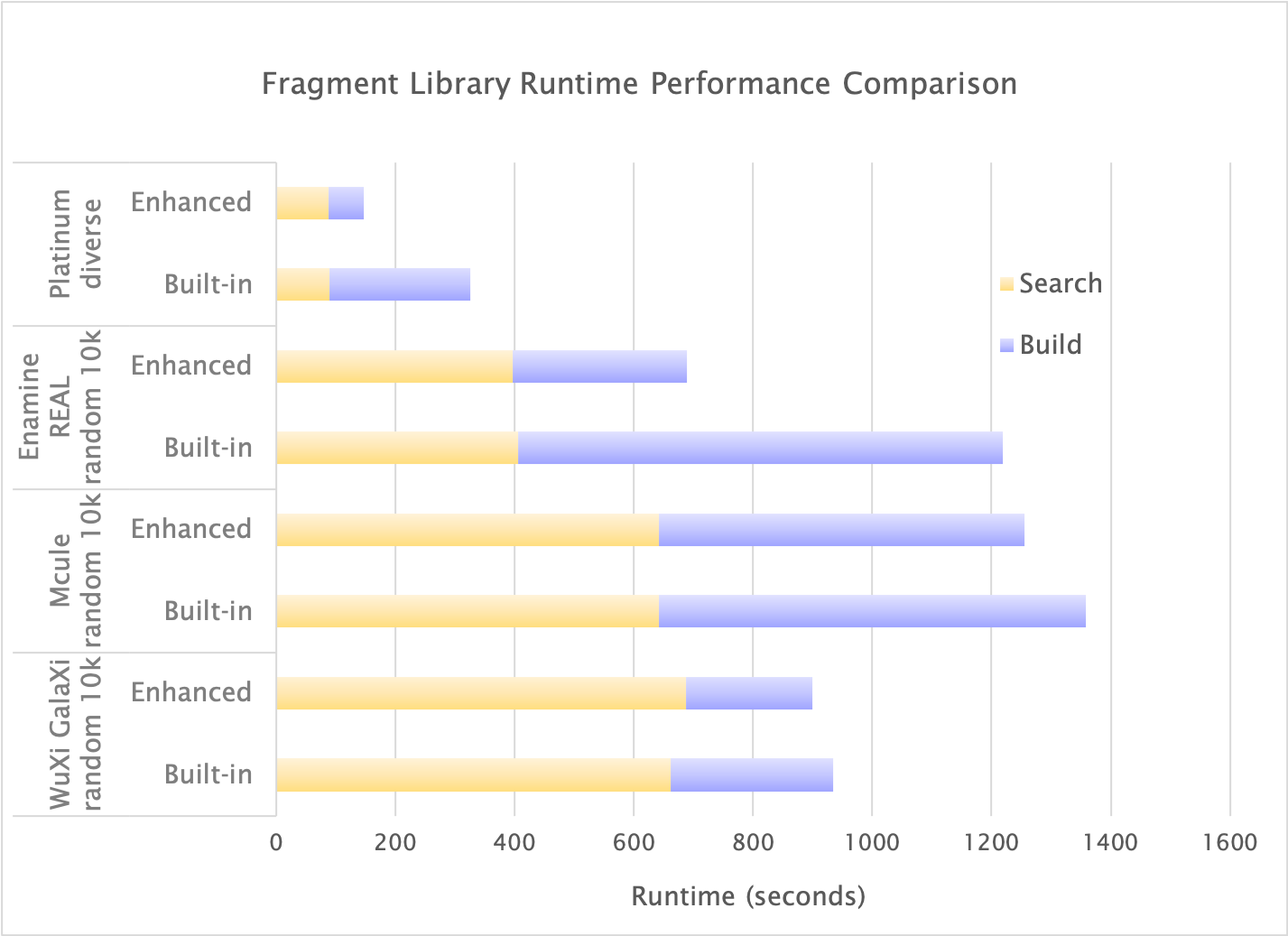
GPU-OMEGA runtime performance using the built-in fragment library compared to the new enhanced fragment library. Benchmark datasets are a filtered set of 2359 molecules of the Platinum dataset and 10,000 randomly chosen molecules of the Enamine Real, Mcule, and WuXi databases.
The enhanced fragment library generates the same high-quality conformers as the built-in library, as shown by the minimum RMSDs of the filtered Platinum dataset.
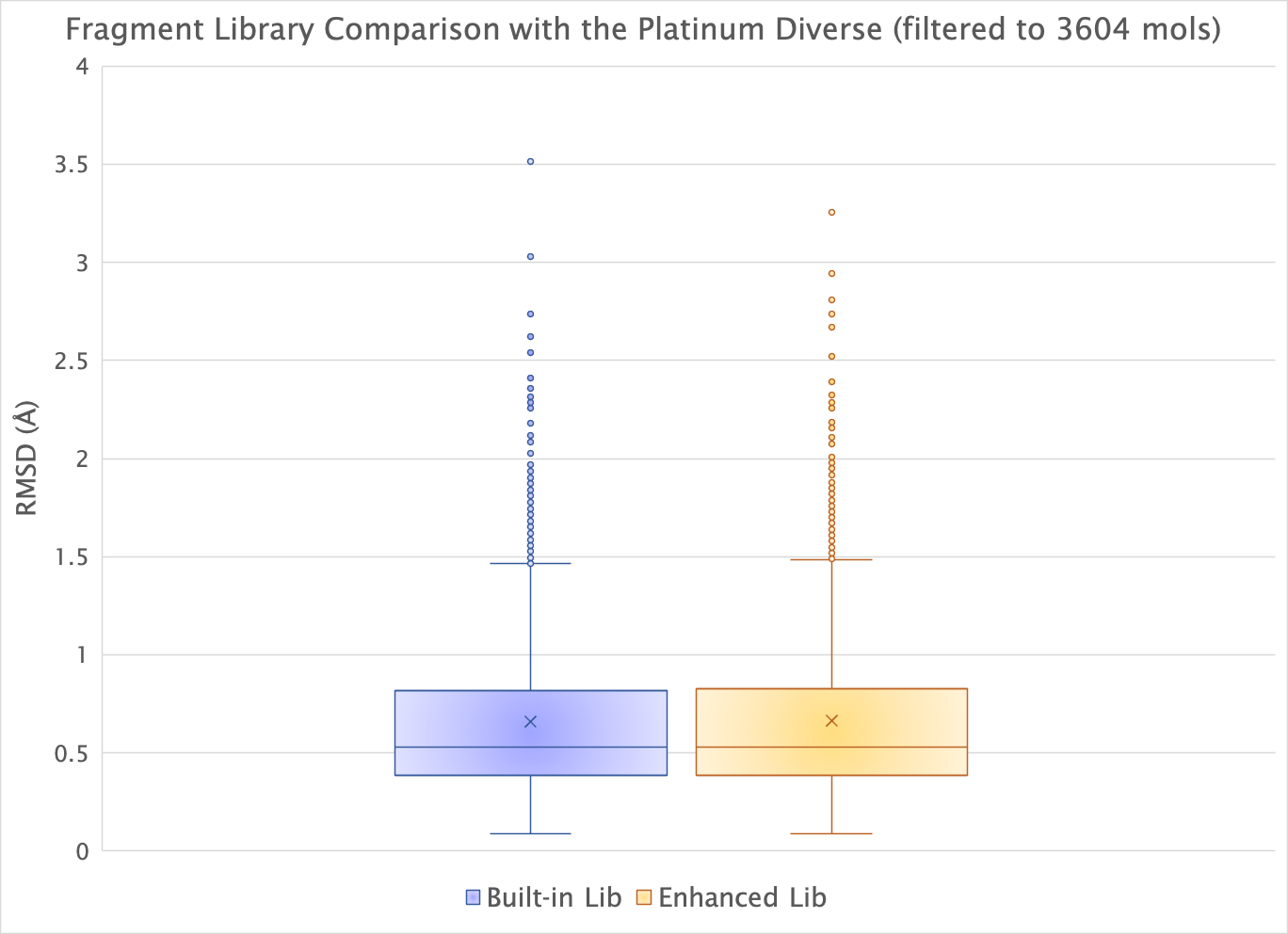
Box plots showing minimum RMSDs of a filtered set of 2359 molecules of the Platinum dataset with the built-in fragment library and the new enhanced fragment library.
- Datasets can be downloaded from:
We are eager to get your feedback on the new enhanced fragment library. Please share your experience with us at support@eyesopen.com.
OEDOCKING: Improved receptors
Receptors used in the OEDocking TK and the OEDOCKING applications,
including FRED, HYBRID and POSIT, have been improved
to take advantage of properly prepared structures from SPRUCE. A new
OEReceptor object has been introduced and is created and contained within an
OEDesignUnit. Since an OEReceptor is now an integral part of an
OEDesignUnit, it is saved into an OEDU file along with a design unit.
There is no separate I/O for the receptors.
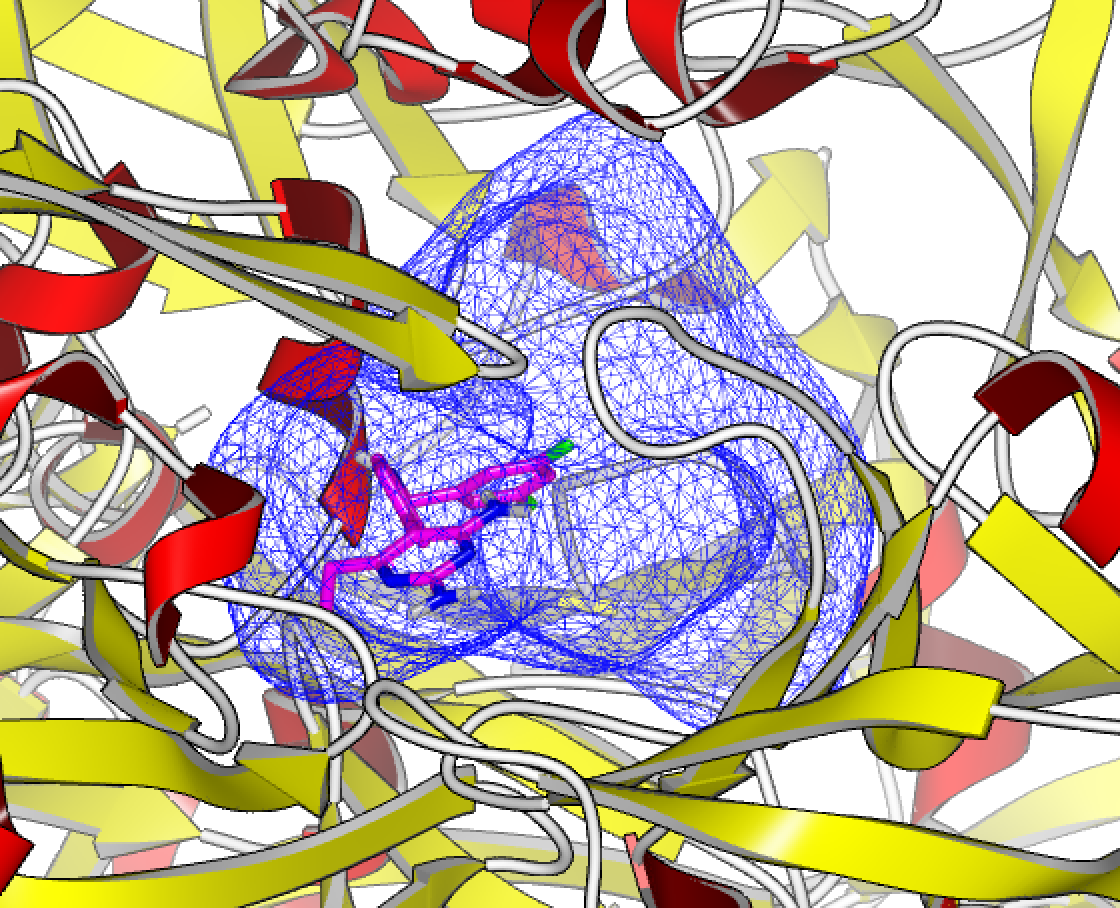
OEDocking receptor, along with the target structure and the bound ligand, for the 2IKO Human Renin Complexed with Inhibitor.
Having the receptors as part of the design unit with properly prepped structures
makes it easier for the docked and posed structures to be used in further downstream
modeling, which is especially necessary with protein force fields like FF14SB. The
new receptor also assists with more flexible use of modern force fields like FF14SB
and Parsley in POSIT.
SZYBKI: A new protein force field
The AMBER FF14SB protein force field has been implemented in OEFF TK. This
new force field has also been made available in SZYBKI for optimizing both
protein and protein-ligand complexes.
FF14SB is the most widely used protein force field for molecular dynamics or
any other force field-based calculations. It is also used in Orion molecular dynamics
package, and is the community gold standard for such calculations.
Supported Platforms
OS
Versions
Linux
RHEL7/8, Ubuntu18/20
Windows
Win10
macOS
10.13, 10.14, 10.15
General Notices
Support for Ubuntu16 has been dropped. Support for Ubuntu20 has been added.