Running Gameplan
The application GamePlan uses SZMAP to analyze ways to modify ligand chemistry based on water structure and energetics in the immediate environment of the ligand. By testing a limited number of coordinates based on the structure of the ligand and the binding site, GamePlan quickly generates: a description of how compatible the ligand is with the binding site, a series of hypotheses for ligand variants, and stabilizing or destabilizing waters.
Again, assuming the input protein and ligand structures have been prepared as described above, a GamePlan calculation can be performed as follows:
> gameplan -szmap_mpi_np 4 -prefix 4std_gameplan -p 4STD_ABC__DU__BFS_A-173.oedu
or
> gameplan -szmap_mpi_np 4 -prefix 4std_gameplan -p 4STD_A__DU__BFS_A-173_target.oeb.gz -l 4STD_A__DU__BFS_A-173_ligand.oeb.gz
The protein and ligand in the OEDesignUnit are analyzed and the most informative sample points are identified. SZMAP is then run on those points, and the results are transformed into a set of potential ligand modifications. All of this takes about 4 minutes on four 2.8GHz processors and the output is placed in the file “4std_gameplan.oeb.gz”, which can be analyzed in VIDA using the WaterColor VIDA Extension. The file “4std_gameplan.log” documents the results, including the energy values for each attachment hypothesis.
The command line option -szmap_mpi_np # controls how GamePlan
runs SZMAP to compute the results. More information on
options can be found in chapter GamePlan.
Analyzing Gameplan Results
Results from GamePlan contain ideas for ligand modifications and points where water is stabilizing or destabilizing the protein/ligand complex.
> vida 4std_gameplan.oeb.gz
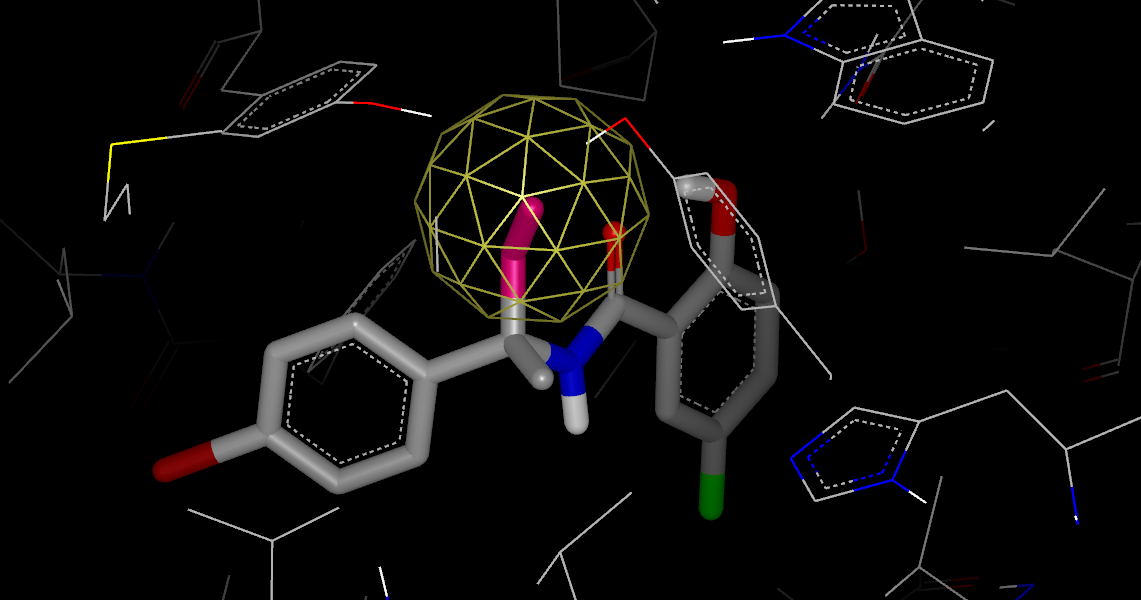
Gameplan Hypothesis for a Polar Attachment
In Gameplan Hypothesis for a Polar Attachment, the WaterColor VIDA Extension has been used to configure the display of the GamePlan results by highlighting any part of the gameplan results in the list window and running WaterColor VIDA Extension. Using the up-arrow and down-arrow keys, you can browse through the different components of the results in the list window. Shown in pink is a suggestion for a site where a hydrogen-bond acceptor might interact favorably with nearby tyrosine residues. The yellow ball indicates that water makes a strong polar interaction at this site in the apo-protein. For a complete description, see chapter GamePlan.