combine_receptors
Overview
In many cases ligands from different receptors may be combined to yield more productive posing targets. For example, if, when the two proteins are aligned, two ligands overlap but occupy different spaces in the binding pocket, combine_receptors will make a single receptor using both ligands that may be a better target for predicting some ligands.
When given multiple receptors, combine_receptors will find the best matched pair for each input receptor. The best matched pair is the combination of two ligands that overlap but have a large difference in shape.
For each set of combined ligands, two receptors are output, one for each input protein. This also helps when analyzing clashes.
Output names are automatically generated so that if two receptors are
merged, say A_receptor.oeb.gz and B_receptor.oeb.gz, two outputs are
formed:
combine_receptors_A_receptor_merged_B_receptor.oeb.gz
combine_receptors_B_receptor_merged_A_receptor.oeb.gz
For example, consider the merged receptors shown in figures Merged receptors and Matched to merged receptor.
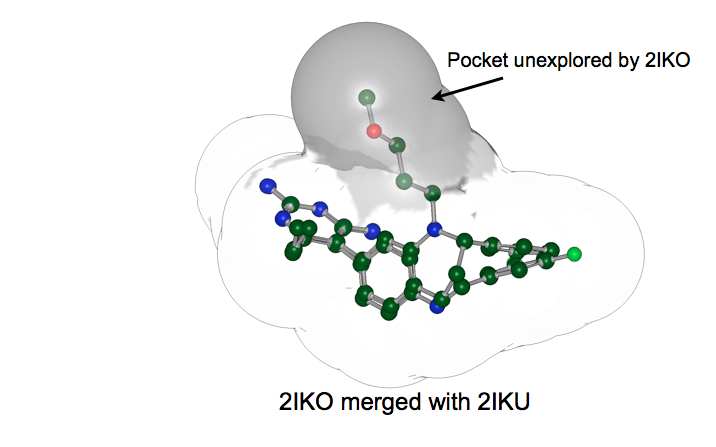
Merged Receptors: Merging the receptors 2IKO and 2IKU capture more potential interaction constraints than either does alone
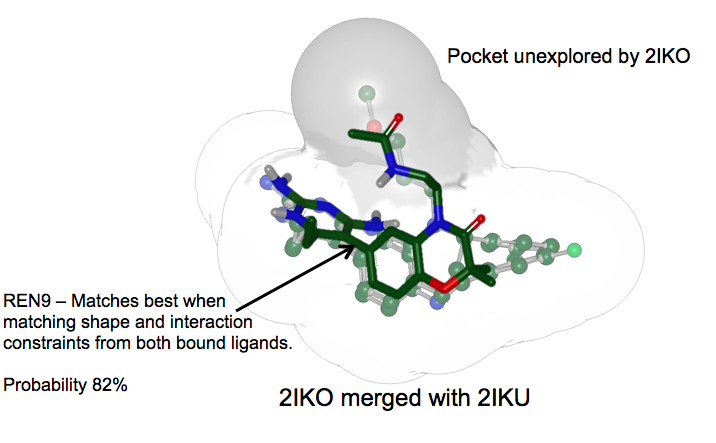
Matched to Merged Receptors: REN9 is correctly predicted by the merged receptor
If a combined ligand has significant clashes with the protein
a warning will also be output with the recommended setting when
running posit.
Serious clashes will not be output unless the user specifies the
-allowedClashes option.
Example Commands
To run combine_receptors, simply provide a list of known receptors:
First, make some receptors:
> spruce -in renin/2IKO.pdb.gz
> spruce -in renin/2IKU.pdb.gz
To convert output OEDesignUnit files into OEBinary files:
> du2oebreceptor -in rec_2IKO_ABCDEFsym1__DU__7IG_A-601.oedu -out rec_2IKO_ABCDEFsym1__DU__7IG_A-601.oeb.gz
> du2oebreceptor -in rec_2IKO_ABCDEFsym2__DU__7IG_A-601.oedu -out rec_2IKO_ABCDEFsym2__DU__7IG_A-601.oeb.gz
...
> du2oebreceptor -in rec_2IKU_ABCDEF__DU__LIY_A-336.oedu -out rec_2IKU_ABCDEF__DU__LIY_A-336.oeb.gz
> du2oebreceptor -in rec_2IKU_ABCDEF__DU__LIY_B-336.oedu -out rec_2IKU_ABCDEF__DU__LIY_B-336.oeb.gz
...
> combine_receptors -receptors rec_*.oeb.gz
Note
On Microsoft Windows systems, you need to expand the wildcard:
> combine_receptors -receptors rec_2IKO_ABCDEFsym1__DU__7IG_A-601.oeb.gz rec_2IKO_ABCDEFsym2__DU__7IG_A-601.oeb.gz rec_2IKO_ABCDEFsym3__DU__7IG_A-601.oeb.gz rec_2IKU_ABCDEF__DU__LIY_A-336.oeb.gz rec_2IKU_ABCDEF__DU__LIY_B-336.oeb.gz rec_2IKU_ABCDEF__DU__LIY_C-336.oeb.gz rec_2IKU_ABCDEF__DU__LIY_D-336.oeb.gz rec_2IKU_ABCDEF__DU__LIY_E-336.oeb.gz rec_2IKU_ABCDEF__DU__LIY_F-336.oeb.gz
The merged receptor files will be named combine_receptors_rec_*:
Wrote combined receptor:combine_receptors_rec_2IKU_ABCDEF__DU__LIY_E-336_merged_rec_2IKO_ABCDEFsym1__DU__7IG_A-601.oeb.gz
Wrote combined receptor:combine_receptors_rec_2IKO_ABCDEFsym2__DU__7IG_A-601_merged_rec_2IKU_ABCDEF__DU__LIY_E-336.oeb.gz ShapeTanimoto of merged ligands:0.690121
...
It is often useful to first collect the receptors ready for combining into a directory, and place the merged receptors
into another directory which must already exist, to do this, simply use the -outputdir option.
> combine_receptors -receptors receptors/*.oeb.gz -outputdir merged
Note
Again, on Microsoft Windows systems, you need to expand the wildcard (this will be omitted for all future examples).
If a merged ligand has an unallowable clash with a protein, it will not result in a receptor.
Command Line Help
A description of the command line interface can be obtained by executing CombineReceptors with the –help option.
> combine_receptors --help
will generate the following output (omitting the OpenEye banner for space)
Help functions:
combine_receptors --help simple : Get a list of simple parameters
combine_receptors --help all : Get a complete list of parameters
combine_receptors --help defaults : List the defaults for all parameters
combine_receptors --help <parameter> : Get detailed help on a parameter
combine_receptors --help html : Create an html help file for this program
combine_receptors --help versions : List the toolkits and versions used in the application
Required Parameters
- -receptors <filenames>
List of receptors to combine. If a file is not a receptor, combine_receptors will halt.
Optional Parameters
- -allowedClashes
Clashes allowed between the ligand and protein of a receptor. There are three levels:
Allowed Clash
Description
noclashes
No clashes are allowed. Actually there is a little wiggle room here less than 0.2 Ångström penetration is not considered a clash.
mildclashes
Mild clashes are allowed (greater than 0.2 Ångström and less than 0.65 Ångström)
allclashes
All clashes are allowed.
The clash ranges have been tuned to account for average coordinate error in a sampling of PDB files, they are intended to be used as guidelines and may not be indicative of some clash states.
By default, receptors are not output if serious clashes exist between the specified ligand and protein. By specifying
-allowedClashes allclashesall clashing receptors will be output.[default = mildclashes]
- -param <filename>
Defines the control parameter file. This file can contain a collection of parameters which can be used instead of writing each parameter to the command-line. In addition, the parameter file written by any posit run (see
-prefixbelow), can be used with the-paramflag in subsequent posit_ executions. Any command given explicitly on the command line will supersede any command found in a file specified with the-paramparameter.
- -prefix
Controls the name of the default param file.
[default = combine_receptors]
- -outputdir
Write the merged receptors to the directory specified (the directory must already exist).
- -verbose
Show that matching status of every receptor pair (whether it is merged or not)
[default = false]