Query Molecule
The upper left contains a depiction of the current query molecule. A query molecule can be loaded by opening a file from the file combo box directly below the depiction, pasting from your favorite molecule editor, or by double clicking the depiction which launches the editor. The editor can accept both common molecule names and SMILES strings.
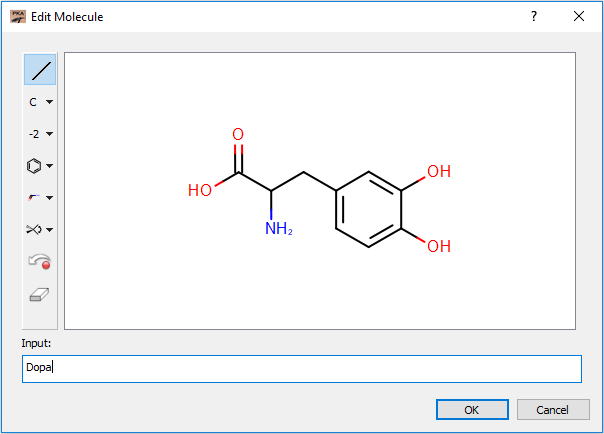
Editor
Once a molecule is loaded the application performs an “Analog Search.” The individual ionizable regions of the query molecule are highlighted. After the “Analog Search” is performed the ionizable regions become clickable, indicated by the highlighting color becoming more saturated when the mouse is hovered above. Clicking on a given ionizable region will limit the results to molecules that match that particular ionizable substructure. Clicking in the negative white space will return the full list of results.
The Query Molecule will persist between sessions.
Tautomers
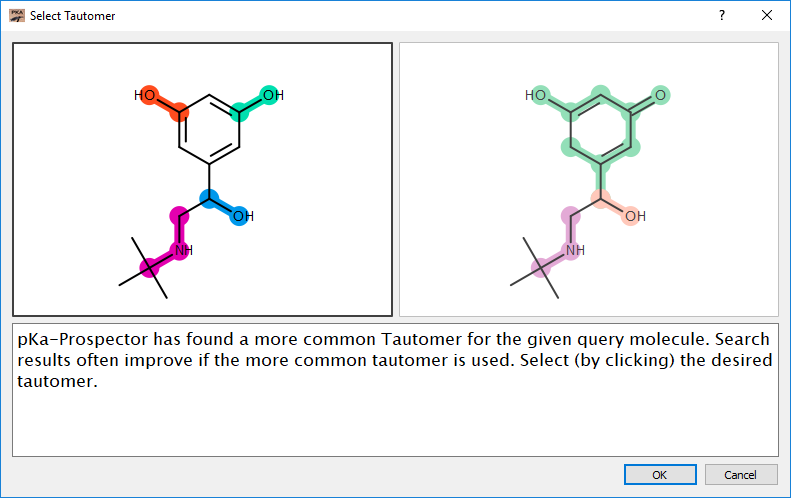
Tautomer Selection
If pKa-Prospector detects a more common tautomer for a given query molecule it will prompt the user to use the more common tautomer. Often search results will improve if the more common tautomer is used as the query molecule.