Setup a simple run and a validation run
vROCS guides the user through all the steps of setting up and performing a ROCS run and visualizing and analyzing the results. There are two main run types for which a user would wish to employ ROCS.
Simple run
Validation run
From the Welcome page click on the button to Perform a simple
ROCS run or Perform a ROCS validation to bring up the Run
set-up dialog.
|
|
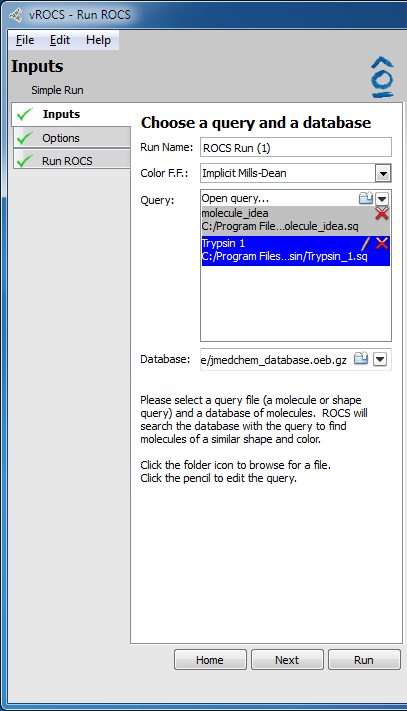
Simple run setup |
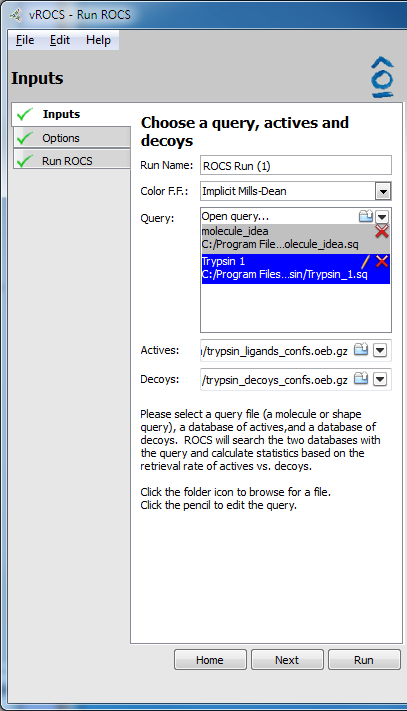
Validation run setup |
- Run Name:
Editable name that will be used for the ROCS run and displaying the results in vROCS.
- Color F.F:
A dropdown menu that allows selection of the current color force field. Options are:
Implicit Mills Dean (default unless changed in User Preferences),
Explicit Mills Dean
If a custom color force field was selected using
ROCS > Preferencesthen this will also be available here. The current force field cannot be changed for a specific active query. Changing the current force field in the dropdown will filter the active query list to show only queries which use that color force field. Opening a saved query file (*.sq, *.sq.gz) will use the color force field previously associated with that file and the active force field in the Color F.F. dropdown will change to reflect this.- Query:
List of queries that can be selected for the ROCS run. Click on
Open queries...or the folder icon to browse to saved ROCS query files (molecules/grids/queries). Click on the black down arrow icon to select from a list of recently used queries. The source file path is shown below the opened query name. Queries built in the vROCS query editor are automatically added to this list for the current vROCS session. The query highlighted in blue is the selected (active) query. The query name can be edited (pencil) or the query can be deleted from the list (red X). Corresponds to the-querycommand line flag.- Database:
The source of ligands which ROCS is to align to the query file during a simple ROCS run. Click on
Open database...or the folder icon to browse to database files. Click on the black down arrow icon to select from a list of recently used databases. Corresponds to the-dbasecommand line flag.- Actives:
The source of ‘active’ ligands which ROCS is to align to the query file during a ROCS validation run. Click on
Open database...or the folder icon to browse to database files. Click on the black down arrow icon to select from a list of recently used databases.- Decoys:
The source of ‘decoy’ ligands which ROCS is to align to the query file during a ROCS validation run. Click on
Open database...or the folder icon to browse to database files. Click on the black down arrow icon to select from a list of recently used databases. The number of decoy ligands cannot exceed 100,000.- Home:
Return to the Welcome screen or the vROCS query editor.
- Next:
Proceed to the Run set-up details Inputs tab. This button only becomes active once the query and database (or actives and decoys) files are selected.
- Run:
Run ROCS using the selected run name, query and database (or actives and decoys) and default parameters. Only becomes active once query and database (or actives and decoys) are selected.
The input for ROCS is a shape-based query with optional color atoms and one (or more) databases of molecules to search. The query shape is most frequently derived from a ligand of interest although other sources are possible, such as a variety of grids built in AFITT, Spicoli, OEDocking, OEChem and third party tools (see ROCS Shape Query Sources). vROCS allows the user to load a pre-saved query for ROCS, having 3D coordinates, or to build or modify one in situ. The list of available queries for a specific run is filtered based upon the type of color force field shown in the Color F.F. drop down. The database(s) are required to be prepared externally with 3D coordinates generated and conformers enumerated, usually by OMEGA. (See Simple run setup and Validation run setup)
Simple Run
A simple run aligns a database of pre-computed molecular conformers against a query. For each molecule in the database it overlays every conformer based on molecular shape with the option to employ color force fields. For a full description of the shape and Gaussian theory employed by ROCS, see Shape Theory. The conformers are scored based upon the Gaussian overlap to the query and the best scoring conformer is reported. The most common scores are ShapeTanimoto (shape alone), or TanimotoCombo (shape + color) which is the default for ROCS and vROCS. The molecules in the database are finally ranked by the scores for their best aligned conformers. This type of simple ROCS run is commonly used when lead-hopping i.e. looking for structurally dissimilar molecules which have a higher probability of biological activity at the same target as the query while also overcoming issues such as ADME/Tox or patent coverage. Numerous literature examples of this application exist and some representative examples are given here.
Note
See List of selected ROCS publications for a list of selected ROCS publications. Lead-hopping examples include:
Validation Run
Before running a simple ROCS run on a large database, e.g. a corporate database of thousands or potentially millions of compounds (and even more conformers!), one should have confidence that the query is indeed able to distinguish true actives from inactives. For this purpose the validation ROCS run is employed. The major difference when setting up a validation run is that the validation run searches two sets of compounds, whereas the simple run searches only a single database. These two datasets are:
A set of molecules known to possess the desired biological activity. These are the actives.
A set of molecules known (or presumed) not to possess the desired biological activity. These are the decoys. The decoys can be a random set of molecules from a database or could be property matched (e.g. DUD [Huang-2006]) for a more stringent validation.
The method of alignment of compounds is the same for both run types (simple and validation). In the case of the validation run the desired result is that molecules from the set of actives are generally scored more highly than the set of decoys i.e. they have a greater shape similarity. Measurement of the degree of selectivity between these two datasets provides the user with confidence that the query is, indeed, selective and suitable for use in a simple ROCS run on a larger dataset.
A good validation experiment is vital to the success of future research. It needs to be carefully planned and set up e.g. selection of active and decoy datasets as well as query design (see Editing ROCS Queries in vROCS). For example, is a modification to a query really beneficial to the selectivity of that query? The rigorous use of validated statistical methods and parameters to analyze and, more importantly, compare runs is essential and frequently overlooked. For that reason statistical analysis tools are included in vROCS when visualizing the results of a ROCS validation run. These are described below in Statistics Metrics.
The run set-up options pages (See Simple run options and Validation run options) in vROCS are pre-populated with the default ROCS options e.g. how compounds are initially oriented and aligned and how alignments are scored and ranked. These default values are calculated to give a good starting point in the majority of examples. However, these are also some of the most common options that a ROCS user might want to modify. For example, changing the start type from inertial to random can be particularly useful for a grid-based query (as opposed to a shape-based query) because it is more difficult to identify and set the 4 true inertial points for a grid. The disadvantage of using random starts and setting this number to be high is that it will significantly increase the run time. Deselecting the 3D view option will speed up runs, particularly on computers with limited compute resources. By default an Open GL 3D alignment for each compound is shown as the run progresses and, since this can be somewhat CPU intensive, switching the display off can be beneficial.
|
|
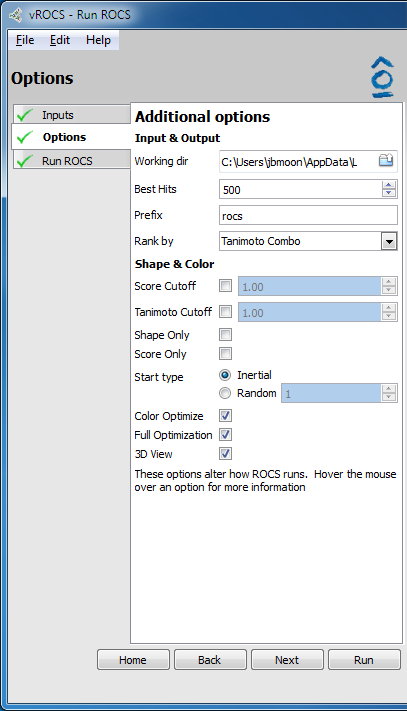
Simple run options |
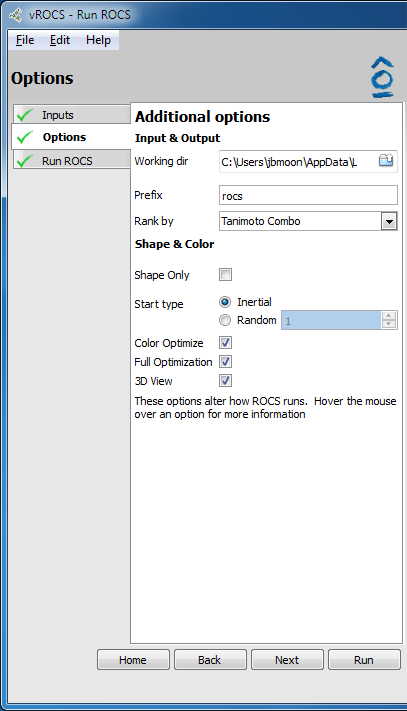
Validation run options |
- Working Directory:
Directory in which the files for the ROCS run are to be saved. Default location is the vROCS installation directory, if it is user writable, otherwise, a temporary directory is used. Click on the folder icon to browse and select alternative directories. Corresponds to the
-outputdircommand line flag.- Best Hits:
Number of top ranking hits to be saved after searching the entire database. Use the arrows to increase/decrease or type the desired number in the field. Corresponds to the
-besthitscommand line flag. Only available for simple ROCS run.- Prefix:
Naming prefix for the current ROCS run. All the output ROCS files will contain this name. If no name is specified the default is “rocs”. Corresponds to the
-prefixcommand line flag. The output files using the prefix are: Parameter file (prefix.parm), Log file (prefix.log), Report file (prefix_1.rpt), Status file (prefix_1.status), Structure file (prefix_hits_1.oeb.gz)- Rank by:
Dropdown allows selection of one of the many score types available in vROCS. The results will be ranked by the selected score for selection of Best N hits (above). Default is TanimotoCombo. Corresponds to the
-rankbycommand line flag. Available scores are: TanimotoCombo, ShapeTanimoto, ColorTanimoto, Combo Reference Tversky, Shape Reference Tversky, Color Reference Tversky, Combo Fit Tversky, Shape Fit Tversky, Color Fit Tversky, Overlap- Score Cutoff:
Check the check box to exclude any hit with a score less than the specified value from the hitlist. The score used is the one specified by the Rank by field. Change the cutoff value by using the arrows to increase/decrease or type the desired number in the field. The allowed score range varies according to the score selected in the Rank by field. Corresponds to the
-cutoffcommand line flag. Only available for simple ROCS run.- Tanimoto Cutoff:
Check the check box to exclude any hit with a ShapeTanimoto score less than the specified value from the hitlist. Change the cutoff value by using the arrows to increase/decrease or type the desired number in the field. The allowed score range is 0-1 (min-max). Corresponds to the
-tanimoto_cutoffcommand line flag. Only available for simple ROCS run.- Shape Only:
Check the check box to perform a shape only overlay, turning off the color force field. Corresponds to the
-shape_onlycommand line flag.- Score Only:
Check the check box to score the incoming poses against the query in their current 3D coordinate frame, turning off alignment and hitlist. This is useful for scoring a pre-aligned dataset. Corresponds to the
-score_onlycommand line flag. Only available for simple ROCS run.- Start Type:
Use the radio buttons to specify how ROCS places the initial alignment. Inertial is the default option and uses 4 initial starts. Random specifies using random starts for the initial overlay and corresponds to the
-randomstartscommand line flag. Specify the number of random starting configurations by using the arrows to increase/decrease or type the desired number in the field.- Color Optimize:
Check the check box to use the color force field in the optimization of the alignments. Default is checked on. Corresponds to the
-optchemcommand line flag.- Full Optimization:
Check the check box to perform full best overlay optimization. Default is checked on. If off (false) then score only. Corresponds to the
-optcommand line flag.- 3D View:
Check the check box to select whether a 3D view of the query and database molecules aligning is displayed as the run progresses. Default is on. If checked off a text-based progress screen is displayed. This will increase ROCS’ run speed on low powered computers.
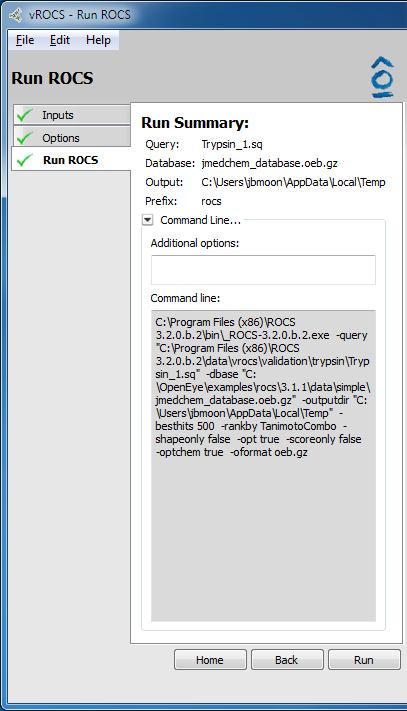
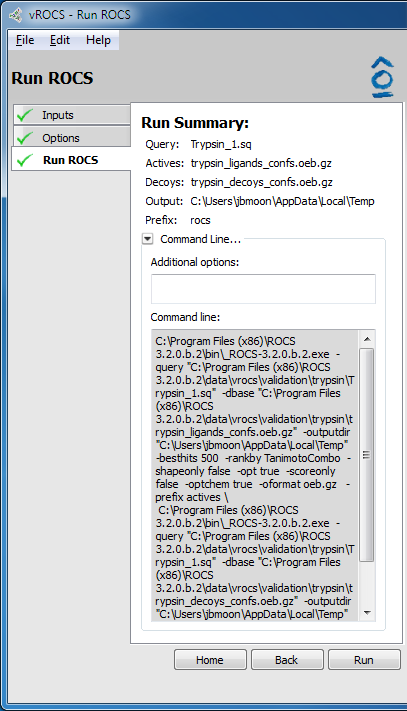
The final page of set-up is the Run Summary on the Run Rocs page. The summary gives a quick rundown of the query file and database used, as well as the ROCS version. It also contains a collapsible panel to display the full set of command line options that will be fed to ROCS and will be saved as the ROCS parameter file (.parm). This can be useful when setting up and validating runs in vROCS that will later be run on the command line across a remote cluster. The Additional Options prompt allows entry of a command not listed in the command line such as a new parameter not yet available in the released version of ROCS.
|
|
Simple run summary |
Validation run summary |
- Query:
Query file as specified on the Inputs tab
- Database:
Database file as specified on the Inputs tab in a simple run
- Actives:
Actives database file as specified on the Inputs tab in a validation run
- Decoys:
Decoys database file as specified on the Inputs tab in a validation run
- Output:
Working directory where all files will be written.
- Prefix:
Naming prefix for the output files that will be written to the Working Directory, as specified in the Options tab. This is defined by the Prefix field on the Options tab.
- Command Line…:
Click to display/hide the full command line that will be sent to the ROCS executable. This can be copied to export and use in command line ROCS installations. A field is available for typing additional ROCS parameters that will be included in the command line not exposed via the vROCS interface. Note that the command line may use temporary files in some instances.