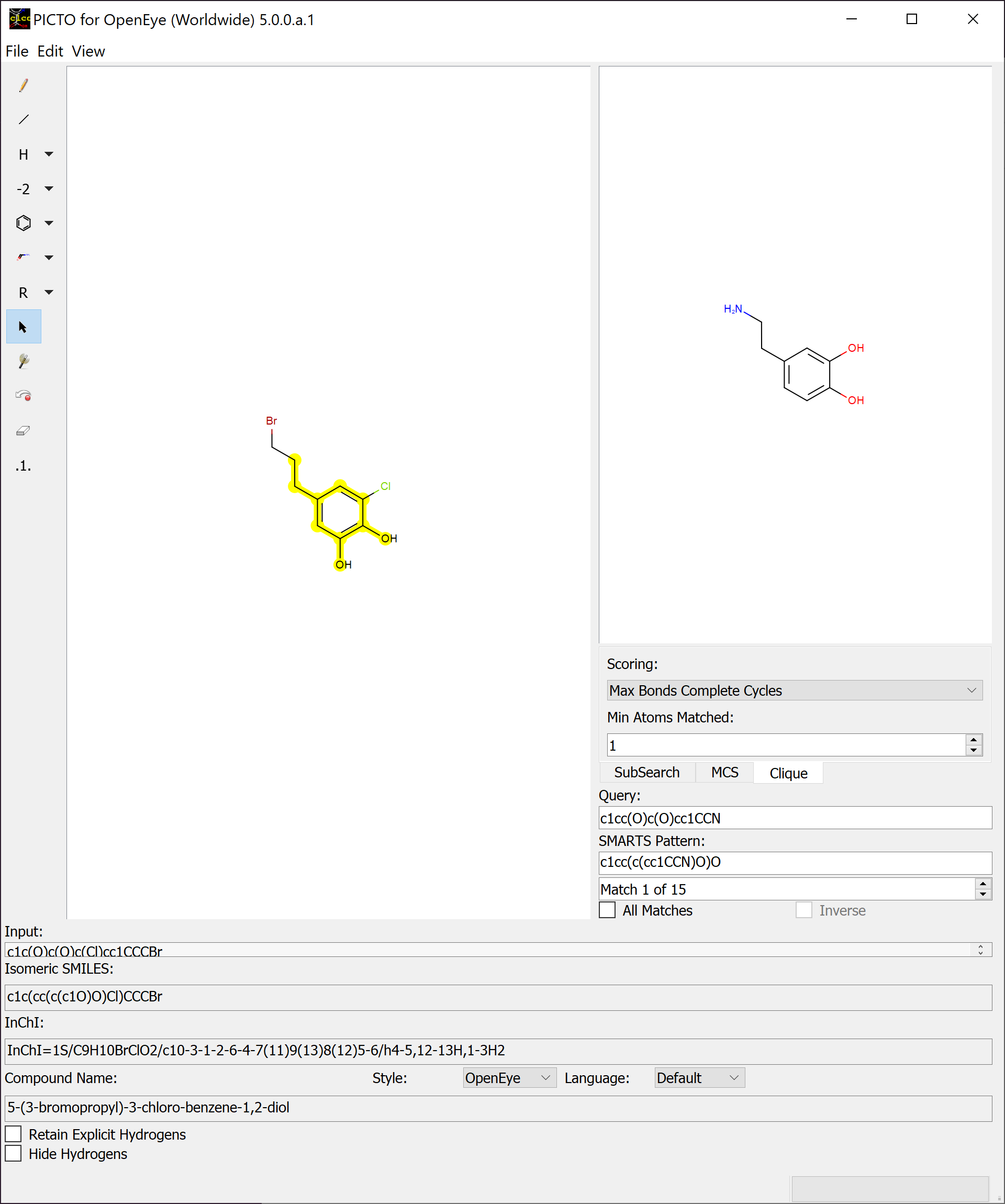
SMARTS¶
The PICTO SMARTS window (populating the right side of PICTO) contains a basic 2D sketching environment, a tab mechanism to cycle through the different pattern matching searches, a SMARTS query entry mechanism, a textbox displaying the SMARTS pattern, a spinbox for matches, a checkbox to display all matches, and an inverse checkbox to see all atoms that did not match. Once a valid SMARTS query is entered, it will be applied to the molecule in the main sketcher and the first match will be highlighted. If an invalid SMARTS query is entered, the text will display red. SMARTS queries can be generated utilizing Send Selected to SMARTS in the Edit tab of the menubar if any portion of a molecule is selected in the main sketcher. The matches can be cycled through using the spinbox and pressing the Enter key after entering a SMARTS query will set focus to the spinbox, allowing the use of the arrow keys to cycle through matches. There are three pattern matching searches available in PICTO: Substructure Search, Maximum Common Substructure Search, and Clique Search.
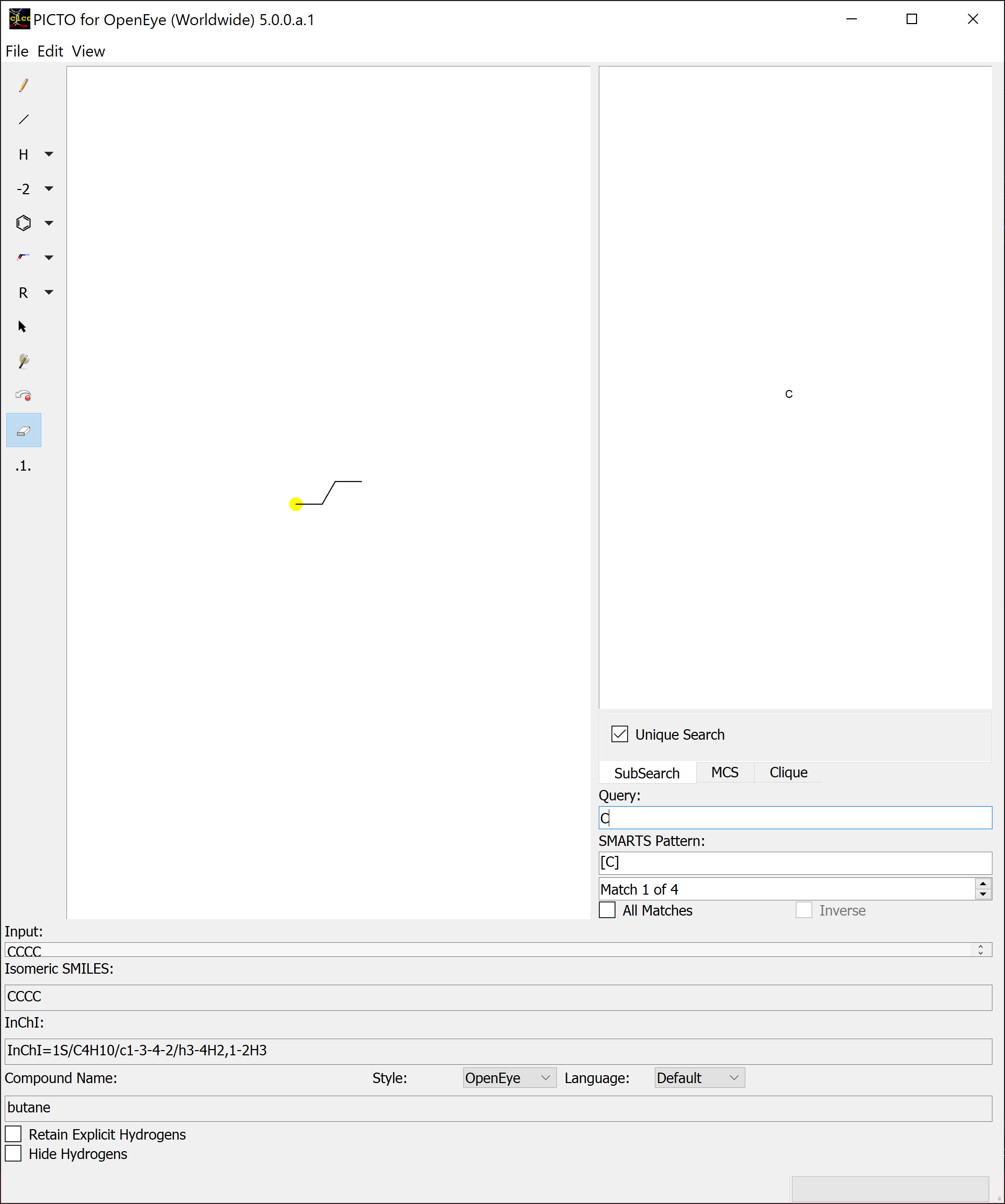
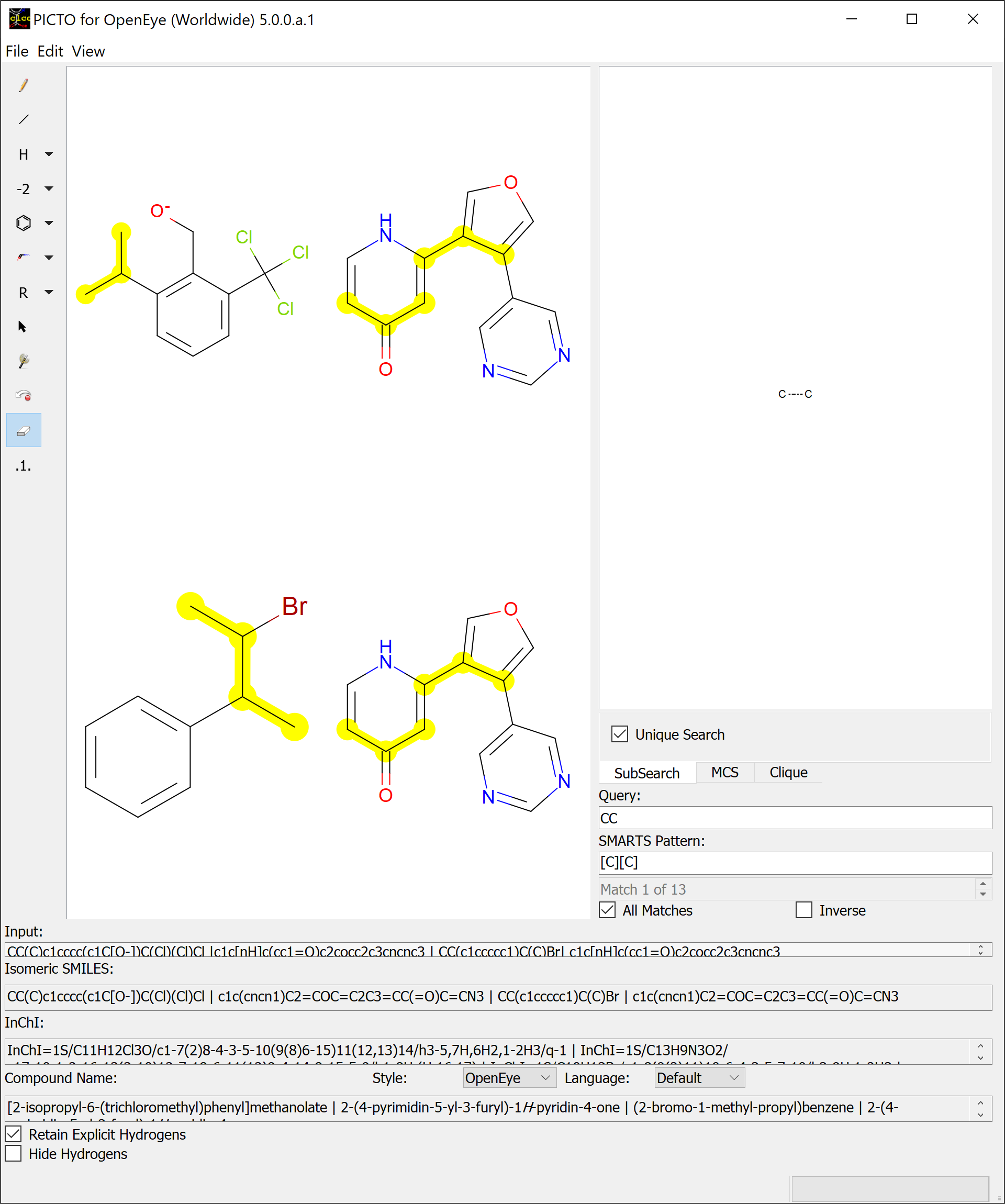
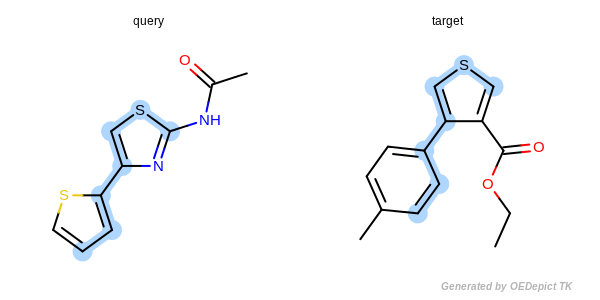
The substructure search (SubSearch) identifies matches by utilizing the query molecule as a submolecule that may be found in the molecules depicted in the main sketcher. A basic overview of a SMARTS query using a basic substructure search can be seen in Figure: SMARTS
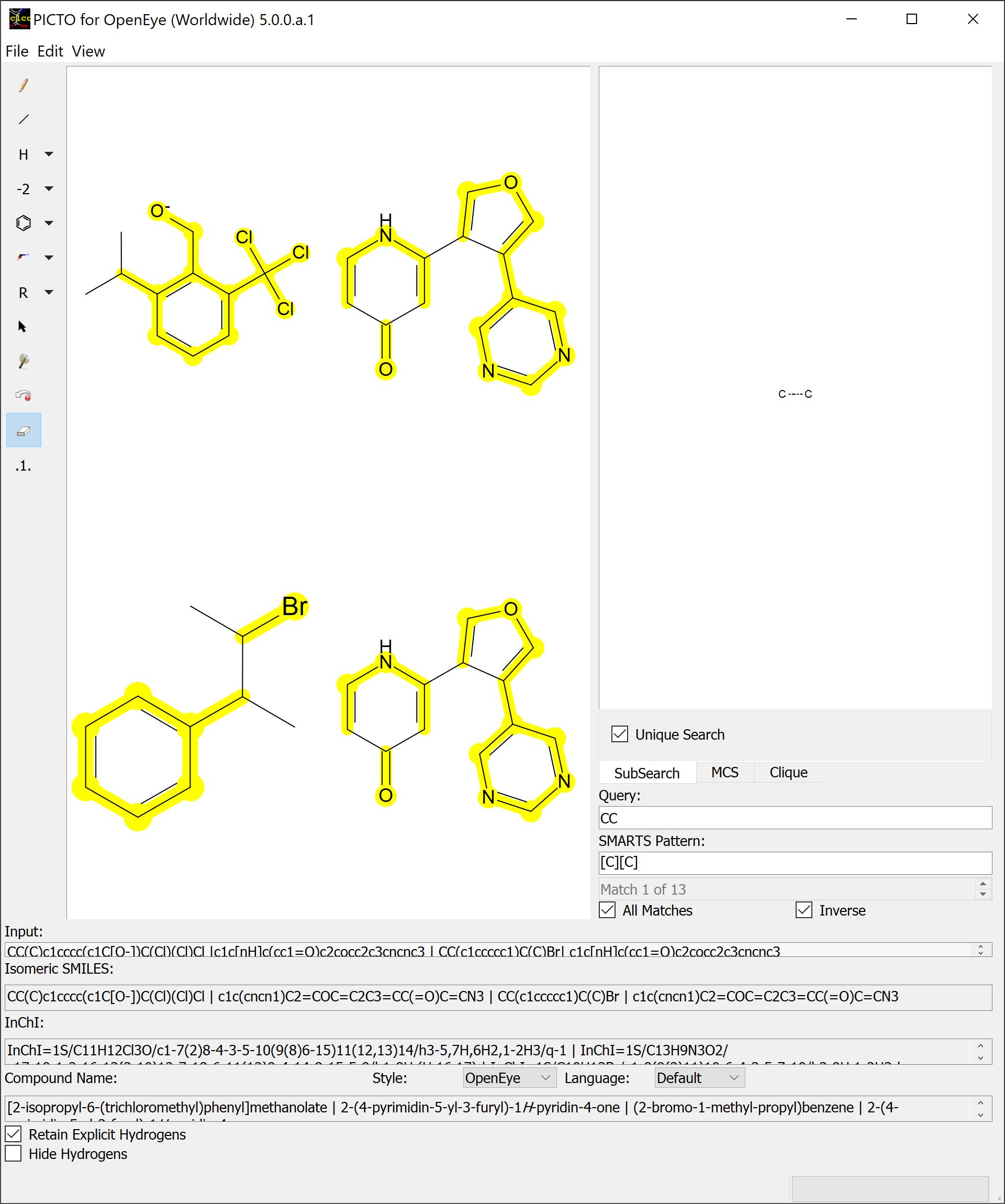
Toggling the All Matches checkbox will highlight every match produced by the search and give access to the Inverse checkbox.
Every atom that didn’t match will be highlighted when the Inverse checkbox is checked.
PICTO defaults to display unique matches, but all matches including duplicates will populate the matches spinbox when unchecking the Unique Search checkbox. The Unique Search checkbox is only available in Substructure Search and Maximum Common Substructure Search.
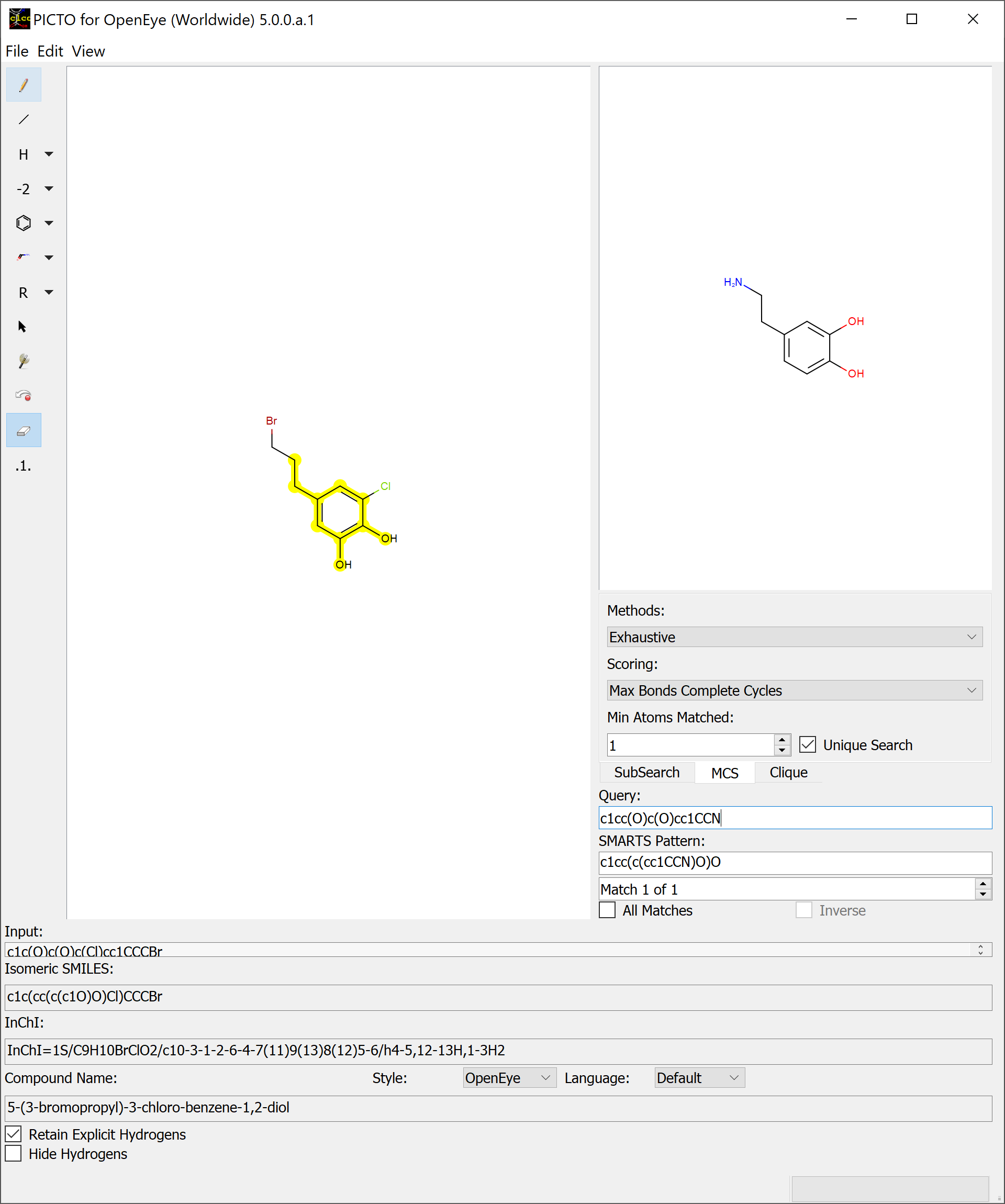
Maximum Common Substructure (MCS) Search identifies the largest match between the query molecule and the molecules depicted in the main sketcher. The MCS tab contains a dropdown for the search method, a dropdown for the scoring method, a spinbox to specify the minimum atoms needed for a match, and the unique search checkbox. The default search method is Exhaustive, which is a more comprehensive method. The other search method available is approximate, which is very fast.
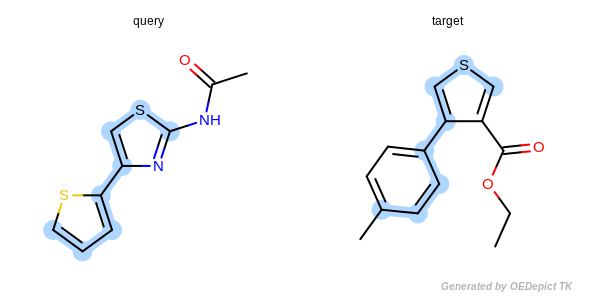
A basic overview of a SMARTS query using a basic maximum common substructure search can be seen in Figure: MCS Search
The approximate method is based on traversing through pre-defined paths of the query structure and trying to map the visited query atoms into target atoms. Because these pre-defined paths represent only a fraction of all possible paths of a compound, it is not guaranteed that the approximate method can find the largest and all common substructures. Significant difference between the detected matches of the two methods could exist in cases when the query or target structure contains complex ring systems (fused or bridged) or stereo centers. However, comparing the two methods for thousands of structures revealed that these cases are rare and the approximate method provides a good trade-off between identifying MCS matches accurately and doing it 3 to 6 times faster than the exhaustive method.
Approximate MCSS¶
Exhaustive MCSS¶
There are four scoring methods available in PICTO are Max Atoms, Max Bonds, Max Atoms Complete Cycle, and Max Bonds Complete Cycle (default).
Max Atoms orders the maximum common substructure matches by the maximum number of atoms included in the graph match. If two matches have the same number of atoms, then the tie is split based on the number of bonds contained in the match.
Max Atoms¶
Max Bonds orders the maximum common substructure matches by the maximum number of bonds included in the graph match. If two matches have the same number of bonds, then the tie is split based on the number of atoms contained in the match.
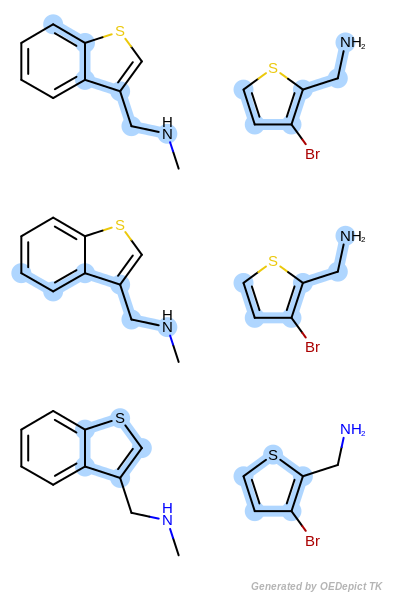
Max Bonds¶
Max Atoms Complete Cycle is the same as Max Atoms with the addition of penalizing cyclic query bonds that are not mapped to any target bonds, thereby giving priority to matches which contain complete cycles common to both the pattern and the target structure.
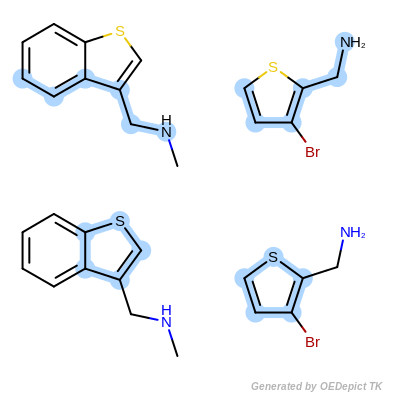
Max Atoms Complete Cycle¶
Max Bonds Complete Cycle is the same as Max Bonds with the addition of penalizing cyclic query bonds that are not mapped to any target bonds, thereby giving priority to matches which contain complete cycles common to both the pattern and the target structure.
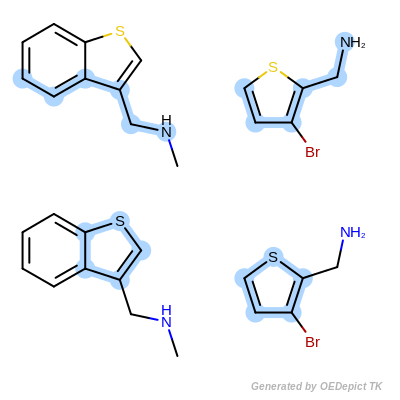
Max Bonds Complete Cycle¶
The Min Atoms Matched spinbox is utilized to add a constraint which sets the minimum number of atoms required of a match to be returned.
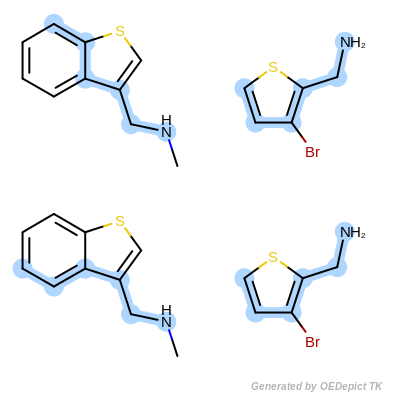
Clique Search (CS) identifies all possible corresponding matches between the query molecule and molecules depicted in the main sketcher. The CS tab contains a dropdown for the scoring method and a spinbox to specify the minimum atoms needed for a match. A basic overview of a SMARTS query using a basic clique search can be seen in Figure: Clique Search. The main difference between this figure and the one above is the amount of matches.