HYBRID tutorial
HYBRID docks molecules using a single receptor or using multiple structures of the target protein. When using a single structure, the input files are simply the receptor and the ligand database. However, when using multiple structures of the target protein, the input files are all of the receptor files and the ligand database.
receptor1.oedu- a receptor file containing the structure of the target protein and a bound ligand.receptor2.oedu- a receptor file containing the structure of the second target protein and a bound ligand. This receptor file should have a different structure of the same target protein inreceptor1.oedu, generally with a different bound ligand.multiconformer_ligands.oeb.gz- conformationally expanded 3D ligands to dock.
See the usage for multiple receptor files in Hybrid Docking with Multiple Crystal Structures.
Setting up and running a HYBRID job is similar to FRED
job. As a comparison to FRED, HYBRID is used to
dock a set of ligands into the same receptor 2IKO_rec.oedu:
> hybrid -receptor renin/receptors/2IKO_rec.oedu -dbase renin/all.oeb.gz
By default, HYBRID generates several output files. Unless you specify a different
Output prefix during setup, HYBRID will use the prefix hybrid for all output files.
hybrid_docked.oeb.gz- top 500 scoring molecules ofall.oeb.gzdocked into2IKO_rec.oedu.hybrid_undocked.oeb.gz- molecules ofall.oeb.gzthat could not be docked into the active site (generally occurs if the molecules are too big for the site). This file will not be present if all molecules were successfully docked to the active site.hybrid_score.txt- a tab separated text file containing the name and score of each of the top 500 ligands.hybrid_report.txt- a text report of the docking process.hybrid_settings.param- a text file containing the parameters used for this run.hybrid_status.txt- a text file that is written periodically during the run with the status of the run.
If you have a multi-processor machine, you can use the OpenMPI option.
> hybrid -mpi_np 4 -receptor renin/receptors/2IKO_rec.oedu -dbase renin/all.oeb.gz -prefix 2IKO_hybrid
As with the output from FRED docking runs, HYBRID results may be viewed and analyzed in VIDA as shown in figure 2IKO HYBRID DOCKED.
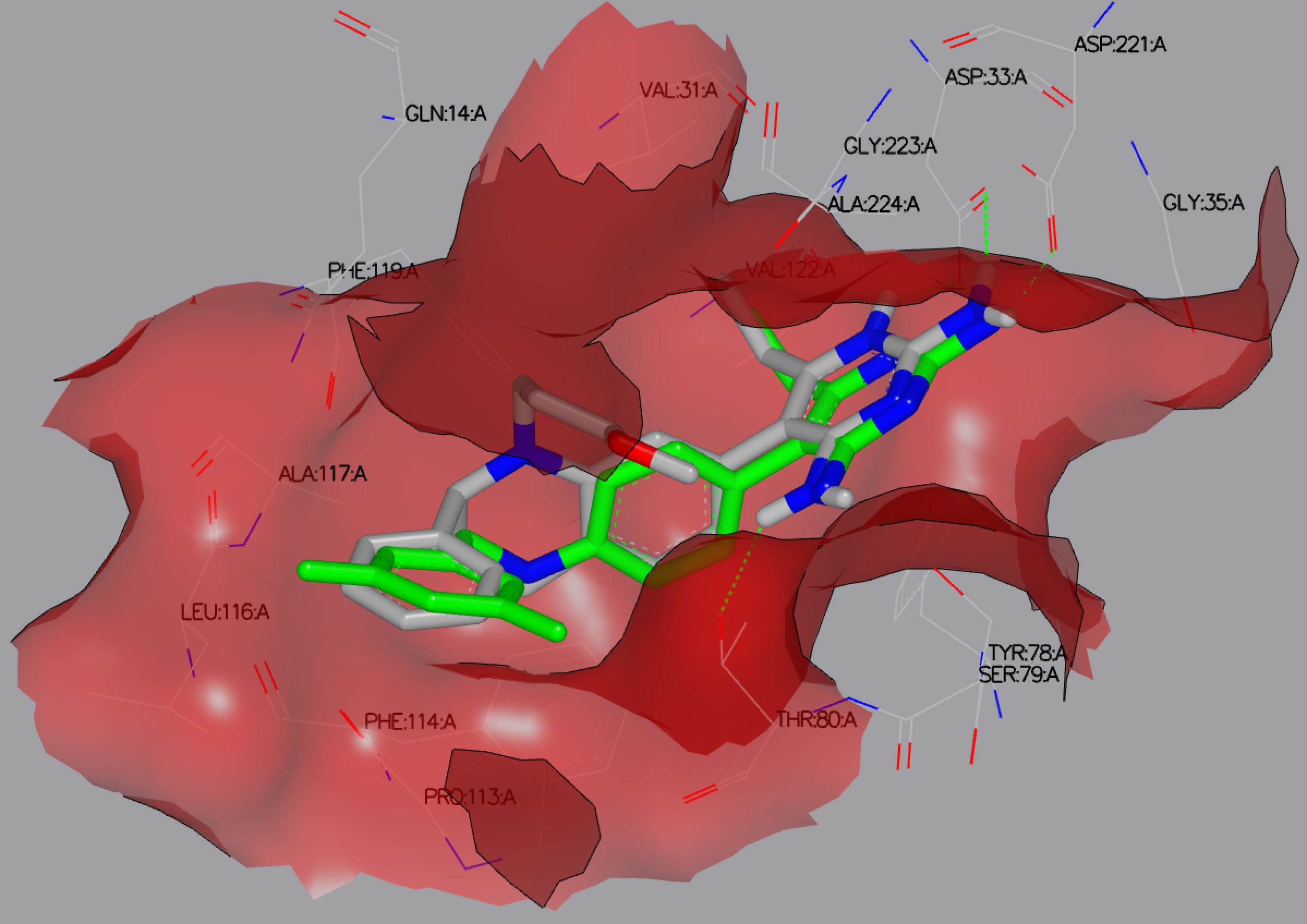
2IKO HYBRID DOCKED results showing the reference ligand (green) and the top docked ligand
Likewise, you also can create a summary PDF report for the HYBRID results using DockingReport.