Example Commands
Running Gameplan
The usual workflow starts with preparing the protein and ligand by adding hydrogens and assigning partial charges. See the SZMAP Workflow for a thorough discussion of preparation.
Next, run GamePlan and view the results in VIDA using the WaterColor VIDA Extension to configure the annotations. On average, a GamePlan calculation will take 4 to 8 minutes on Linux using four 2.8GHz processors, for example.
> gameplan -szmap_mpi_np 4 -prefix 2xdl_gameplan -p 2XDL_AB__DU__2DL_A-1225.oedu
or
> gameplan -szmap_mpi_np 4 -prefix 2xdl_gameplan -p 2XDL_AB__DU__2DL_A-1225_target.oeb.gz -l 2XDL_AB__DU__2DL_A-1225_ligand.oeb.gz
> vida 2xdl_gameplan.oeb.gz
In Gameplan Hypothesis, the bonds in hot-pink represent an idea about how to modify the ligand to add a two-atom hydrogen-bond acceptor group that could interact favorably with nearby tyrosine residues. The yellow ball indicates that water makes a strong polar interaction at this site in the apo-protein.
When GamePlan reads input molecules, it checks for missing hydrogens
and missing partial charges and will halt if the input does not appear to
be complete. The option -warn_if_missing_hydrogens can be used to
prevent halting, but before using it you should analyze the problem to see
if it is due to an error in the input or even a disagreement with OEChem
as to the proper bond order and charge state for your molecule.
Mistakes in encoding alternate conformation codes can, for example, lead to
very long and very short bonds, with the short ones interpreted as double bonds.
And aromatic or double bonds that are not flat enough can cause OEChem to
think they are single bonds. See Protein and Ligand preparation for SZMAP and GamePlan
for discussion of preparation and how to resolve such pathologies using
SPRUCE.
By default, GamePlan hypotheses respect the hybridization of
aromatic atoms in the input ligand. Specifying the option
-sp3_aromatic will permit GamePlan to explore the
effect of a change of hybridization of aromatic atoms.
Note that only the local hybridization at the attachment site is altered,
often leading to odd-looking geometry. But the point is to generate fresh ideas
for how you might modify your ligand.
Gameplan Results
Open the results in VIDA, highlight any part of the output in the List window, and run the WaterColor VIDA Extension to configure the annotation and focus the view to the region around the ligand. See the last section of chapter WaterColor VIDA Extension for a step-by-step guide to doing this.
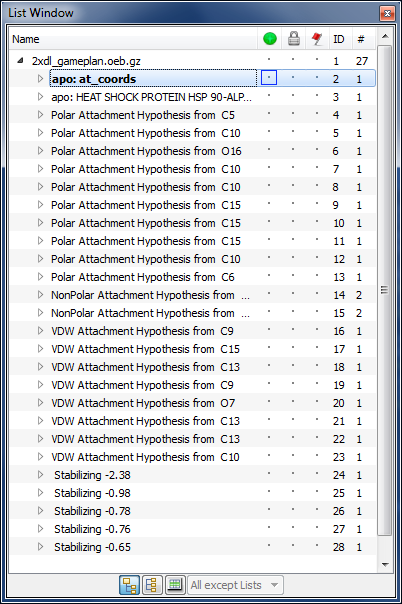
Gameplan List Window
GamePlan results are organized into three main sections (see figure Gameplan List Window):
An analysis of how well the ligand compares to the solvent in the binding site with colored annotation around atoms that indicates the type of site (figure Ligand Compared to apo Binding Site). The protein is also included here for context.
A series of hypotheses for substituents that could be added to the ligand. Each attachment is modeled using dummy atoms that show up in hot-pink in VIDA, with annotation that indicates the type of site: polar, nonpolar and van der Waals (see figures Hypothesis for a Polar Substituent and Hypothesis for a Nonpolar/VDW Substituent).
A (possibly empty) section showing stabilizing and destabilizing water sites (figure Stabilizing Site).
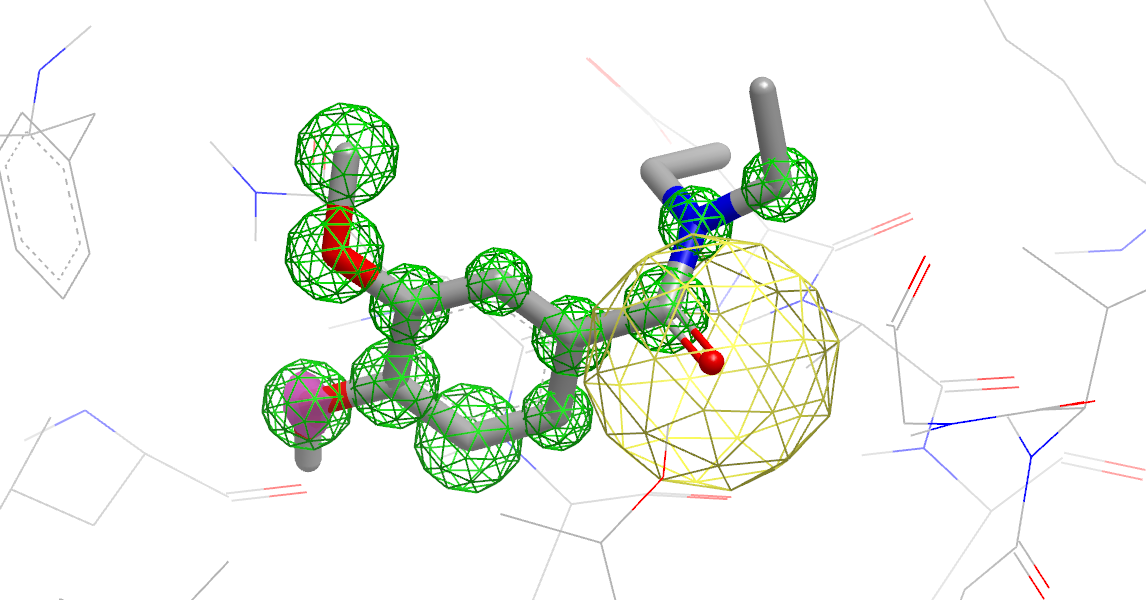
Ligand Compared to apo Binding Site
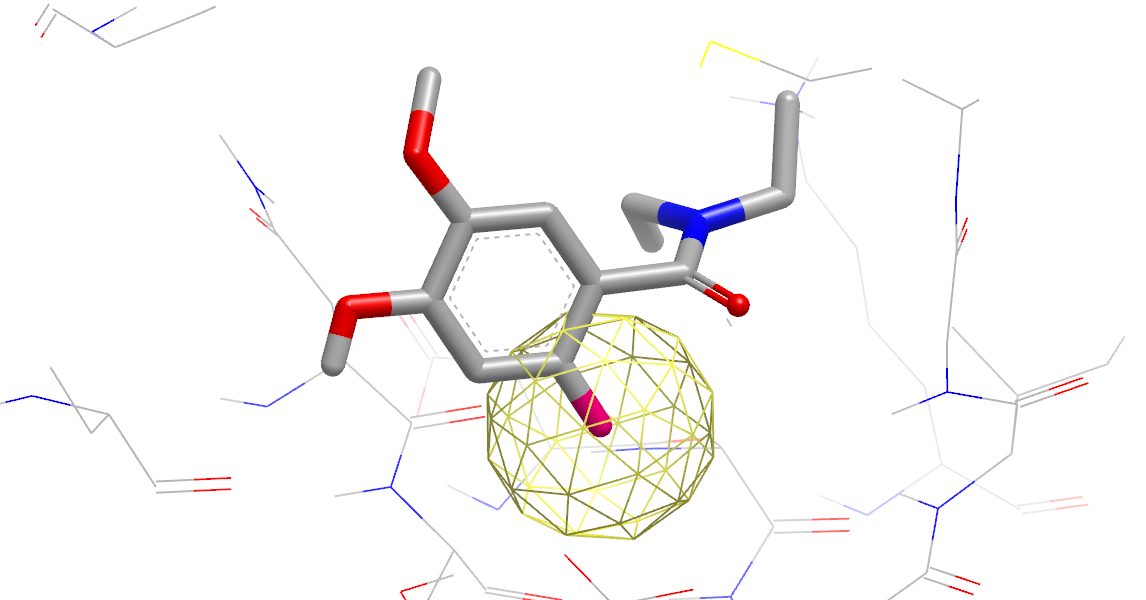
Hypothesis for a Polar Substituent
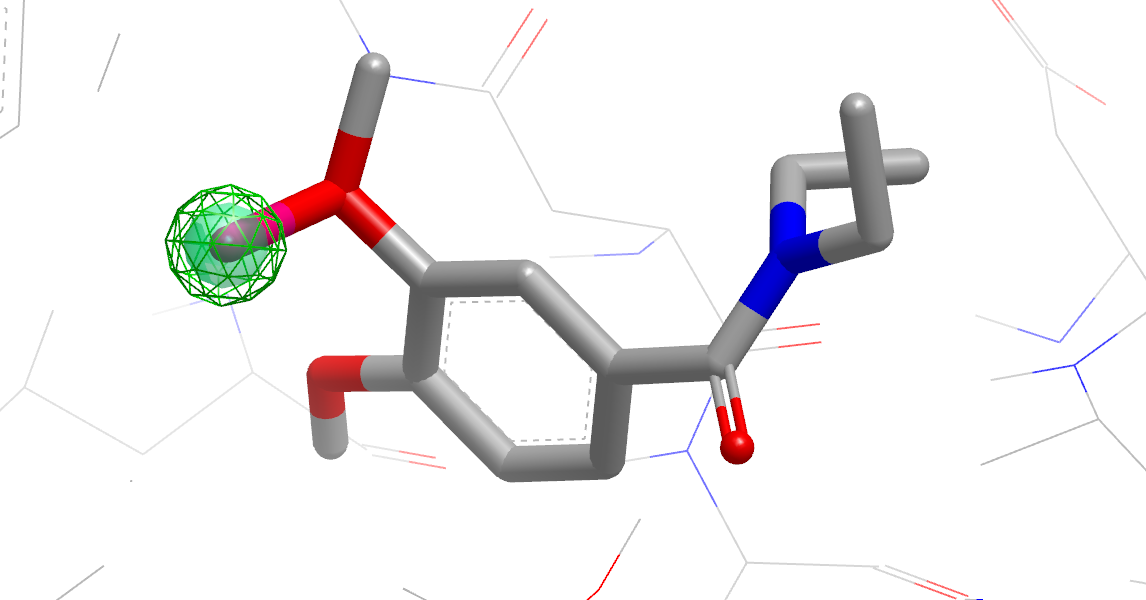
Hypothesis for a Nonpolar/VDW Substituent
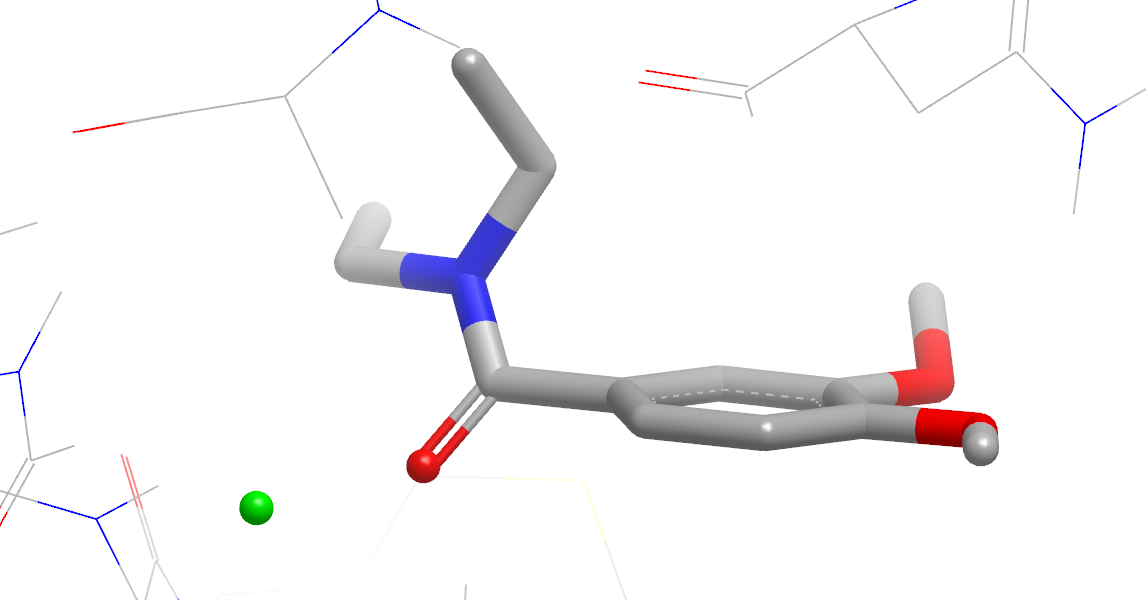
Stabilizing Site
Annotations are:
Yellow mesh sphere: polar site; the size of the sphere indicates the magnitude of the polar interaction.
Green mesh sphere: van der Waals site; the size of the sphere indicates the magnitude of the VDW interaction.
Green translucent sphere: non-polar site.
Purple top: mismatch site, where the ligand polarity does not match that of the solvent.
Small green and red spheres: stabilizing and destabilizing points, respectively.
Based on your results, you may want to modify your ligand to make it more compatible with the apo pocket solvent electrostatics, add a substituent based on the suggested hypotheses, displace a destabilizing water or carefully replace a stabilizing water, mimicking the interactions of the water.