Processes
The Process Manager allows the user to start, monitor, terminate, and load results from processes external to VIDA. As part of this functionality, it is possible to write out currently loaded molecules (or grids) to files which can be used as input to the external processes. Furthermore, it is possible to create “pipes” which chain together the input and output of individual processes into a larger process.
Process Manager
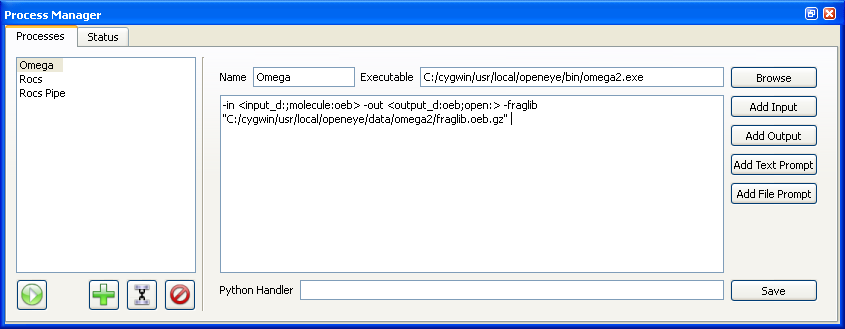
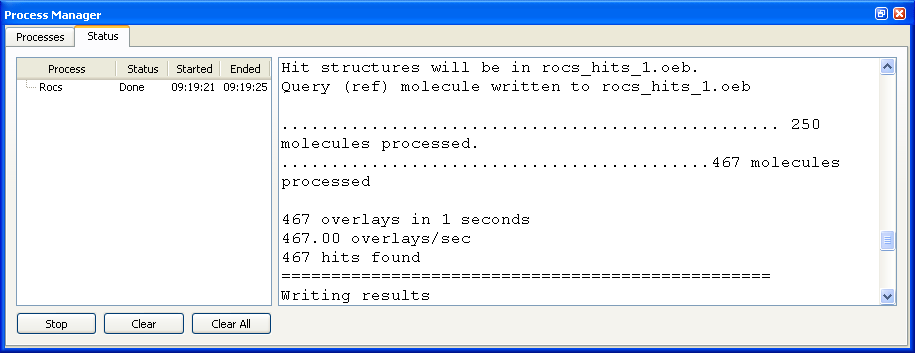
The Process Manager contains two tabbed windows called “Processes” and “Status” respectively as can be seen in Figures X.Y and X.Y. The “Processes” tab contains the list of available processes and the functionality to create and edit them. The “Status” tab displays all of the currently active and recently completed processes and information about their state. It also displays the commandline output generated by each of the processes.
Output generated by a completed process
Beneath the list of processes in the “Processes” tab is a collection of four buttons. The button on the left with the play icon starts the currently selected process. The next button with the plus icon clears the editor on the right hand side to allow for creation of a new process. The next button with the link icon changes the editor on the right hand side to allow for creation of a new pipe (see Figure X.Y). The last button with the delete icon deletes the current selected set of processes from the list of available processes.
Processes
When creating a new process or editing a current one, there are a number of available options. At the top of the editor there is a field for the process name (which will be displayed in the list of available processes seen on the left as described above) as well as a field for the actual location of the application to be run. The “Browse” button to the right of this field can be used to find the location of the desired application.
Beneath this is a large text entry box which allows for specification of any desired command line arguments to the application. It is not necessary to enter the application name here.
For most applications, at least one filename will have to be specified as a command line argument. This can be done multiple ways. If the file to be specified is always going to be located in the same location, the filename can be entered directly into the text entry box as a command line argument. If the location of the file is not likely to be constant, a prompt can be specified as opposed to an actual filename. To add a prompt, click on the “Add File Prompt” button to the right of text entry box. If the desired file will need to be created each time, it is possible to create the desired file at start-time by writing out selected molecules (or grids) directly from VIDA. To use this feature, click on the “Add Input” button to the right of the text entry box. At start-time, the user will be prompted to select the desired molecules (or grids), which will then be saved to a file which will be passed as a command line argument to the process.
In addition to being able to specify specific input files, it is possible to specify an output file which can potentially be read back into VIDA when the process is completed. As discussed above, it is possible to a specify specific filename in the command line or to use a prompt, but if it desired to load the resulting file, the “Add Output” button should be clicked to specify the file.
Besides being able to specify filenames, it is often desirable to be able to specify other variable arguments at start-time. To do this, click on the “Add Text Prompt” button which will add a prompt argument to the command line arguments. At start-time the user will be asked to specify the desired text to be passed to the process.
Lastly, at the bottom of the Process Editor is a “Python Handler” field which allows for specification of a Python function to be run when the process is completed. This allows for handling of output not normally handled by VIDA or for output which is not specified on the command line.
Adding Input
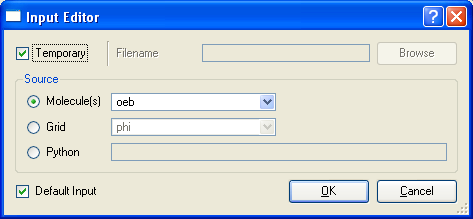
The Input Editor allows the user to specify a mechanism by which an input file is provided to the process to be run. At the top of the editor are two mutually exclusive options, the first is the “Temporary” check box which indicates that the file to be used will be a temporary file which will be removed after the process has completed. The other option is a field which allows specification of a specific filename to be used. In both cases, the user has a choice as to how this file will be populated: saving molecules, saving grids, or running a specified Python function which will handle the population of the file. If molecules or grids are chosen, a file format to save these in must be specified in the pull-down box next to the selected option.
If a Python function is specified to populate the input file, the desired filename can be obtained by using the keyword “@FILE@” in the function specification. This keyword will be substituted with the real filename at start-time before the Python function is called.
Finally, there is a checkbox at the bottom of the dialog to specify whether or not this is a default input. The default input entry is used when constructing pipes.
Process Input Editor
Adding Output
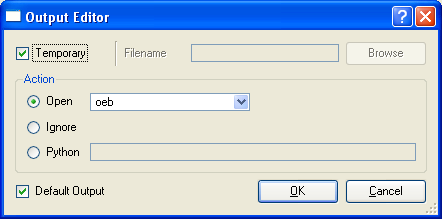
The Output Editor allows the user to specify a mechanism by which an output file of the process to be run is handled by VIDA. At the top of the dialog are two mutually exclusive options, the first is the “Temporary” check box which indicates that the output file is a temporary file and will be removed after it has been handled by VIDA. The other option is a field which allows specification of a specific filename to be used for the output file. If this option is selected, the file will not be deleted after it is handled.
Once the above has been specified, the user can specify how VIDA should handle the file when the process is finished. There are three possible options, the first is to open the file, the second is to ignore the file, and the last is to call a Python function to handle the resulting file. If it is desired to open the file, the file format that the process should use to generate the output file must be specified so that VIDA will know how to properly load the results.
If a Python function is specified to handle the output file, the actual filename can be obtained by using the keyword “@FILE@” in the function specification. This keyword will be substituted with the real filename at process completion time before the Python function is called.
Finally, there is a checkbox at the bottom to specify whether or not this is a default output. The default output entry is used when constructing pipes.
Process Output Editor
Adding Prompts
As described previously there are two types of prompts that are available for specification on the command line: a text prompt and a file prompt.
The Text Prompt Editor (see Figure X.Y(a)) allows the user to add a text prompt to the process as well as to specify what the caption of the prompt will be. Specification of an informative caption is helpful in allowing the user to know what information is being requested.
|
|
Text Prompt Editor |
File Prompt Editor |
The File Prompt Editor (see Figure X.Y(b)) allows the user to add a file prompt to the process as well as to specify what the caption of the resulting prompt will be. Specification of an informative caption is helpful in allowing the user to know what specific file or type of is being requested.
The result of both of these prompts are passed as command line arguments to the process in the order in which they appear in the main text entry box in the Process Editor (see Figure X.Y).
Pipes
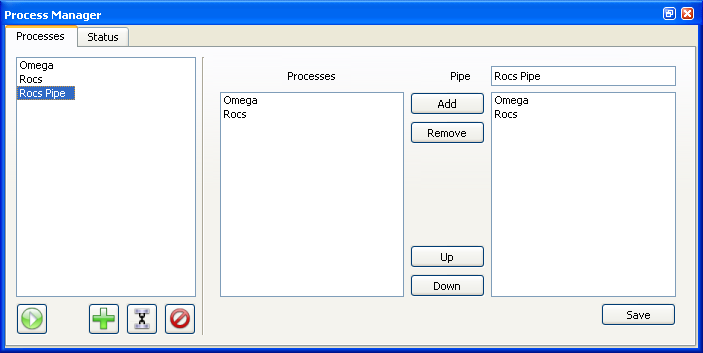
Pipe Editor
Once a number of processes have been created, they can be “piped” together using the Pipe Editor (see Figure X.Y). On the left-hand side of the editor is a list of all the currently defined processes. On the right is an ordered list of processes to be run and in which order to run (top to bottom).
When chaining processes together into a pipe, it is important that they have default input and output fields specified. The default output of a given process is passed into the default input of the following process. When the final process is completed, the user will be notified and the final output file will be handled based on its specification in the associated process.