File I/O
Opening Files
There are multiple mechanisms by which a file can be loaded in VIDA. The simplest method is to select the Open option in the File menu. This will launch a file selection dialog which prompts the user for the desired file(s) which will subsequently be loaded. A short list of the most recently opened files can also be found in the File menu under the Recents (or Open Recents on Mac OS X) submenu. Selecting an option in this submenu will load the associated file. There is an Open All option at the bottom of the submenu. Files can also be loaded by specifying them on the command line when starting VIDA, by dragging the file of interest onto the VIDA desktop icon, or by dragging the file of interest into the List Window.
In addition to the Open menu item, there is a separate Open State item which filters out all files except for state files. State files can be opened using any of the mechanisms described above, but their special nature warrants its own option.
There is also an Open Special submenu which contains three default options: Script, From PDB, and MTZ. The Script option prompts for a file, but filters out all files except for Python scripts. The From PDB option prompts for a PDB ID and then fetches the associated molecule directly from the PDB. The MTZ option prompts for a crystallographic MTZ file, and then allows the user to select the maps and fields to be loaded from the file. The Open Special submenu also serves as an excellent target area for adding custom open operations via the scripting interface.
Molecules
Multiple molecular file formats are supported for reading and include :
OpenEye binary format (version2: .oeb)
OpenEye design unit format (.oedu)
MDL, RDF, and SDF
Tripos Mol2, Mol2H
Daylight SMILES, canonical SMILES, and isomeric SMILES
ChemDraw CDX
ISIS Sketch
MacroModel
MOPAC
PDB, MMCIF
XYZ
As is often the case with file formats, the meaning and use of certain fields within a format may change over time which can potentially lead to problems interpreting those files. VIDA, using OpenEye’s OEChem toolkit, makes its best effort to interpret all files correctly; however, it does provide a mechanism to allow the user to override the handling of certain formats (SMILES, PDB, Mol2, XYZ, and MacroModel). The ability to change the flavor of a specific format can be done in the application preferences. Figure: Molecule flavors.
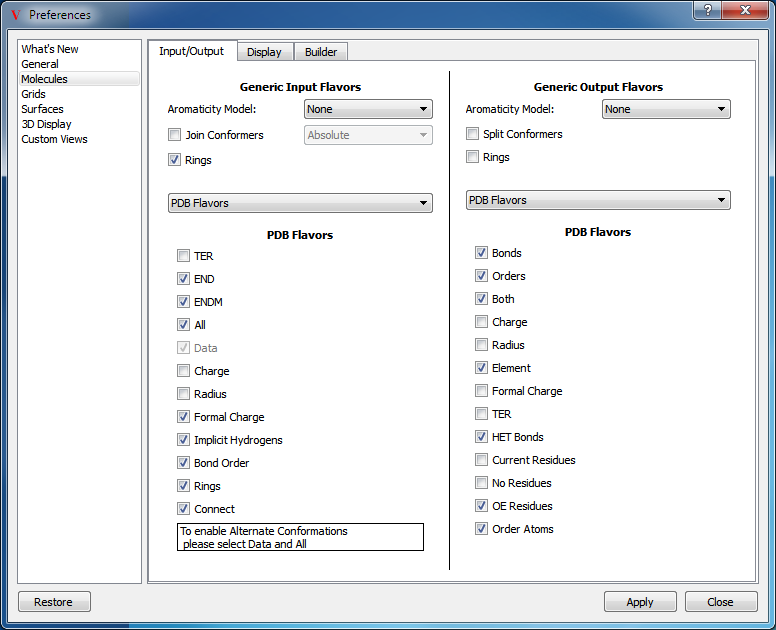
Molecule flavors
An important flavor to be aware of is the Data flavor of the PDB format. Enabling this flavor allows VIDA to properly handle alternate residue locations specified in the PDB file.
In addition to changing the flavor of a format, there are a few other advanced options available when reading molecules including aromaticity model specification and conformer joining. The desired aromaticity model to be applied can be specified in the pull down menu next to the Aromatic Model label. The Join Conformers checkbox controls whether or not adjacent molecules in an input file will be tested on reading to determine whether they are unique compounds or simply different conformers of the same molecule. The specific test to be performed can be specified in the pull down menu next to the checkbox. For more specific details on the available aromaticity models and conformer tests, please see the OEChem documentation.
Note
Because these advanced options are available through the application preferences, they will be remembered by VIDA and automatically applied in future Open operations unless they are subsequently modified or restored.
Grids
Multiple grid and map formats are supported for reading and include:
OpenEye grids
ASCII grids
GRASP/DelPhi grids
CCP4 maps
XPLOR maps
MTZ maps
MTZ maps are a little special in the sense that the contents of the files do not follow a standard format and the relevant data columns for reading phases and amplitudes can be named arbitrarily. By default, VIDA attempts to find common column names that are used by standard refinement packages.
If a MTZ file cannot be opened, an error message is displayed suggesting the use of the menu item File/Open Special/MTZ…
To have more control over which maps are created, use the File menu option Open Special/MTZ…. This opens a dialog that allows selection of both the MTZ columns and the desired output maps.
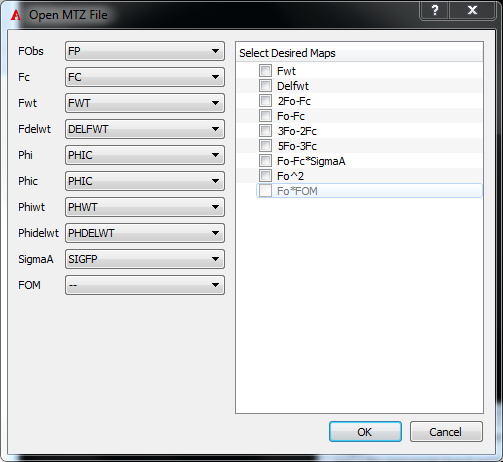
MTZ Open Dialog
Simply select the appropriate columns and map types to generate and click “OK”.
Surfaces
Multiple surface formats are supported for reading and include:
OpenEye surfaces
GRASP surfaces
State Files
The entire state of a session can be stored in a single file called a State File (.oes). A state file contains all of the molecules, grids, and surfaces that were loaded in VIDA at the time the state file was created. In addition, the state file preserves the actual view, layout, and properties of that session. State files provide an extremely convenient way to save sessions for later work or to share with collaborators. Furthermore, state files are the fastest method of reading and writing large data sets.
To load a state file, choose the Open State option in the File menu and then select the desired state file. State files can also be loaded from the command line. It is important to note that loading a state file during a run will clear the current state before loading the new one. Therefore, be sure to save the current state before loading a new state if keeping the current state is desired. Furthermore, since state files contain the entire state of the application that generated them, loading a state file will overwrite local preferences; however, they will not be saved on exit.
Note
State files are not backwards compatible with older versions of VIDA
Python Scripts
VIDA supports reading of Python scripts compatible with Python version 3.7+. VIDA supports two additional file extensions for Python scripts (.pyv and .vpy) to enable users to associate those file extensions with VIDA without adversely affecting the association of unrelated Python scripts.
For more details about Python scripting in VIDA, see the chapter on Scripting as well as the associated API manual.
Closing Files
When a file is opened in VIDA, its contents are loaded into its own individual list. To close a given file, simply right-click on the associated list in the List Window and select the Delete option. This will close the file and remove the contents from the application. However, if an object is present in multiple lists, deleting one of those lists will not delete that object from the other lists. It is important to note that deleting a list, does not affect the actual file that was read.
To close all of the currently loaded files, select the Clear All option in the File menu. In addition to clearing everything that is loaded, this option effectively restores the state of the application to what it was when the application was started.
Saving Files
VIDA is capable of saving multiple types of files including molecules, grids, surfaces, and state files. The details of the individual formats can be found in the relevant sections below.
Note
VIDA does not currently support writing to oedu file format
Prompt dialog
A file can be saved by choosing the Save option in the File menu. This will launch a dialog prompting the user to select which objects to save (see Figure: Prompt Dialog). There are two sets of buttons at the top of this dialog which provide mechanisms for filtering the available options. The three buttons on the top left toggle the display of molecules, surfaces, and grids respectively. The two buttons on the top right provides filters for showing only the Marked and/or Visible objects respectively. Once one or more objects have been selected and the OK button is pressed, a file dialog will appear allowing the user to specify the desired file. The file formats listed in the file dialog filter will reflect which file formats are supported based on the objects selected. Only the OpenEye binary format supports writing multiple object types (molecules, grids, and/or surfaces) to the same file.
This process can be expedited by simply right-clicking on the desired objects and selecting the Save option. This allows the user to bypass the selection dialog and go directly to the file dialog.
Molecules
Multiple molecular file formats are supported for writing and include:
OpenEye binary format v2 (.oeb)
MDL, RDF, and SDF
Tripos Mol2, Mol2H
Daylight SMILES, canonical SMILES, and isomeric SMILES
ChemDraw CDX
MacroModel
Molecular Formula (MF)
MOPAC
FASTA
PDB
XYZ
As is often the case with file formats, the meaning and use of certain fields within a format may change over time which can potentially lead to problems interpreting those files. VIDA, using OpenEye’s OEChem toolkit, writes all files according to the standard; however, it does provide a mechanism to allow the user to override the handling of certain formats (SMILES, SDF/MDL, PDB, MOL2, MF, MOPAC, and MacroModel). The ability to change the “flavor” of a specific format can be done in the application preferences as seen in Figure: Molecule flavors.
In addition to changing the flavor of a format, there are a few other advanced options available when writing molecules including aromaticity model specification and conformer splitting. The desired aromaticity model to be applied can be specified in the pull down menu next to the Aromaticity Model label. For more specific details on the available aromaticity models, please see the OEChem documentation.
Note
Because these advanced options are available through the application preferences, they will be remembered by the application and automatically applied in future Save operations unless they are subsequently modified or restored.
Grids
Multiple grid formats are supported for writing and include:
OpenEye grids
ASCII grids
GRASP/DelPhi grids
Surfaces
Multiple surface formats are supported for writing and include:
OpenEye surfaces
GRASP surfaces
State Files
The entire state of a session can be stored in a single file called a State File (.oes). A state file contains all of the molecules, design units, grids, and surfaces that were loaded in VIDA at the time the state file was created. In addition, the state file preserves the actual view, layout, and properties of that session. State files provide an extremely convenient way to save sessions for later work or to share with collaborators. Furthermore, state files are the fastest method of reading and writing large data sets.
Note
State files are not backwards compatible with older versions of VIDA
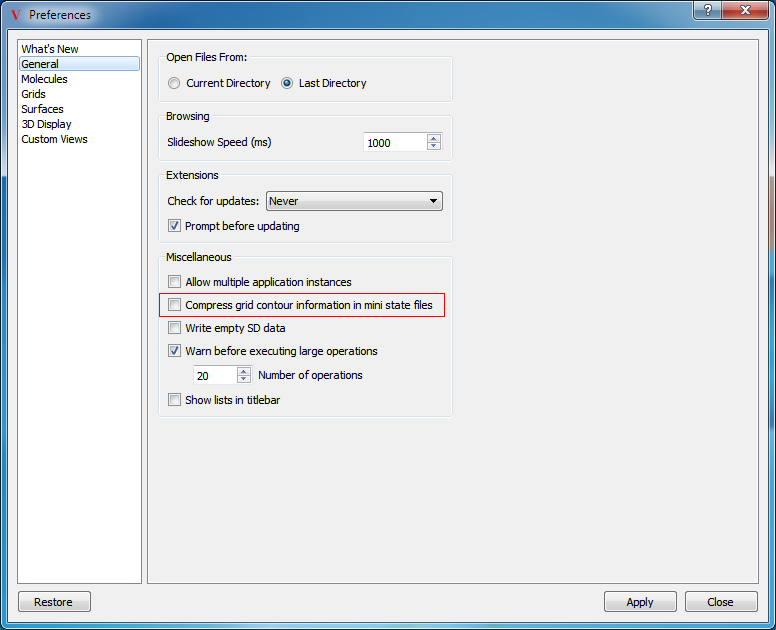
Compress Grid Contours
VIDA supports a “mini” state file option which only writes out objects that are currently visible or referenced in the display bookmarks. This can provide for substantial savings in disk size and load time. This mini-state can be further compressed by selecting the “Compress grid contour information” option in the General section of the application preferences Figure: Compress Grid Contours. Selecting this option zeroes all non-contour related positions in the grid which greatly improves its compressibility. This should only be done if there is no expectation that the user loading the resulting state file will want to look at grid contour levels other than those already defined when the file was generated.
Saving State
To save a state file, select the Save State option in the File menu and specify the desired state file. The state file type can be selected using the file format filter as seen in Figure: Saving State.
Importing Files
External spreadsheet data can be imported into VIDA and associated with currently loaded objects. Both comma separated files (.csv) and tab delimited files (.txt) are supported by default. Once a file has been selected, an import dialog will appear which allows the user to customize the column delimiters, column headers, as well as the criteria on which rows are matched to currently loaded objects. For more details on importing spreadsheet data, please see the Spreadsheet chapter.
Exporting Files
Multiple special file types can be exported from VIDA including data, images, and scripts. Exact details about these files can be found in the relevant sections below.
Data
Spreadsheet data can be exported in three formats: comma separated files (.csv) , tab delimited files (.txt), and PDF (.pdf). Data can be exported by selecting the “Spreadsheet” option in the Export submenu in the top-level File menu. Selecting this option will launch a file dialog which allows the user to specify the output file. Once the desired file has been chosen, another dialog will appear which allows the user to select which spreadsheet and which columns in that spreadsheet should be exported. For more specific details on exporting spreadsheet data please see the Spreadsheet chapter.
Images
POV-Ray
The current scene in the 3D display can be exported to a POV-Ray input file which can be used to generate very high quality and resolution images. To export a POV-Ray file, select the POV-Ray option in the Export submenu in the File menu. The application will then prompt the user to specify a file to which the scene will be saved.
Python
A complete history of the application’s operations can be written out as a Python script. This script can be exported by selecting the Script History option in the Export submenu in the File menu.
Drag and Drop
A powerful drag and drop interface is provided for passing molecules, images of molecules, and molecular data between applications. Multiple mechanisms of transferring data via the drag and drop interface are supported.
Dragging a list of filenames into the List Window will cause those files to be loaded.
Dragging a list of SMILES into the List Window will cause those SMILES to be parsed into new molecules and stored in a new list called “Pasted”. These new molecules will not be assigned 3D coordinates and therefore will not be visible in the 3D display.
Dragging a list of IUPAC or common chemical names into the List Window will cause those names to be parsed into new molecules (using OpenEye’s Lexichem toolkit) and stored in a new listed called “Pasted”. Please note, this functionality requires a separate license for the Lexichem toolkit available from OpenEye. These new molecules will not be assigned 3D coordinates and therefore will not be visible in the 3D display.
Dragging molecules from other molecular visualization programs into the List Window will cause those molecules to be loaded into a new list called “Pasted”. The ability to load molecules from other applications depends on how the other applications place molecules on the clipboard. Currently supported clipboard formats include SMILES, ChemDraw CDX, ISIS Sketch, MDL SD Files, and OpenEye OEB files. Furthermore, for other molecular visualization programs that support a drag and drop interface, molecules can be dragged from the List Window into those applications.
Dragging molecules from the List Window into other applications will cause molecules, molecular data, or molecular images to be transferred to that application depending on how that application expects to receive data. For instance, dragging molecules into a text editor will enter a list of SMILES corresponding to the dragged molecules.
Dragging a 2D depiction from the Spreadsheet will cause that image to be loaded in the associated application if possible.
Dragging molecules within the List Window can be used to organize files and molecules. See the List Management section for more details.
DesignUnits cannot be modified with Drag-and-Drop at this time
Copy and Paste
A copy and paste interface exists for passing molecules, images of molecules, and molecular data between applications. The Copy option can be found in the Edit menu and can also be performed by pressing Ctrl+C on the keyboard. This will copy all of the molecules in the current scope to the clipboard, unless the active window is the Spreadsheet in which case it will copy all of the selected Spreadsheet data to the clipboard instead.
The Paste option can be found in the Edit menu and can also be performed by pressing Ctrl+V on the keyboard. This will paste any molecules found on the clipboard into a new list called “Pasted”.