Release Highlights 2019.Nov
SPRUCE: Application Suite for Biomolecular Structure Preparation
The 2019.Nov release introduces SPRUCE which takes experimental
biomolecular structure data in either PDB or mmCIF formats, and prepares the structures
for use with downstream modeling applications.
With SPRUCE, the output for the prepared proteins is the recently
developed OEDesignUnit object serialized to a new OEDU (OpenEye Design Unit) file format.
The prepared system is componentized into molecular categories including protein, ligand,
nucleic, solvent, and many other molecular types common to biomolecular experiments.
By default, hydrogens are added to systems using a tautomer search for any ligand, cofactor or other nonstandard residue molecules.
In addition, the system is split into distinct biological units and all final design unit structures
are superposed into a common frame of reference.
SPRUCE has been tested across a broad range of industry-relevant targets. It is built using the recently released Spruce TK, which has been used for virtual screening and molecular dynamics simulations in Orion, as well as to prepare structures for MacroMolecular Data Service (MMDS).
The figures below show one of the prepared OEDesignUnits for the PDB code 1TGV. 1TGV is a structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate. In the figure on the left the entire homo-pentamer is shown. The binding site residues and ligand atoms are shown explicitly with a wireframe representation to highlight the site of interest. Five OEDesignUnits are generated for this PDB code, one focusing on each of the 5 binding sites in the pentamer. The binding site view shows the ligand bound pose with OEInteractionHints highlighting various protein-ligand interactions.
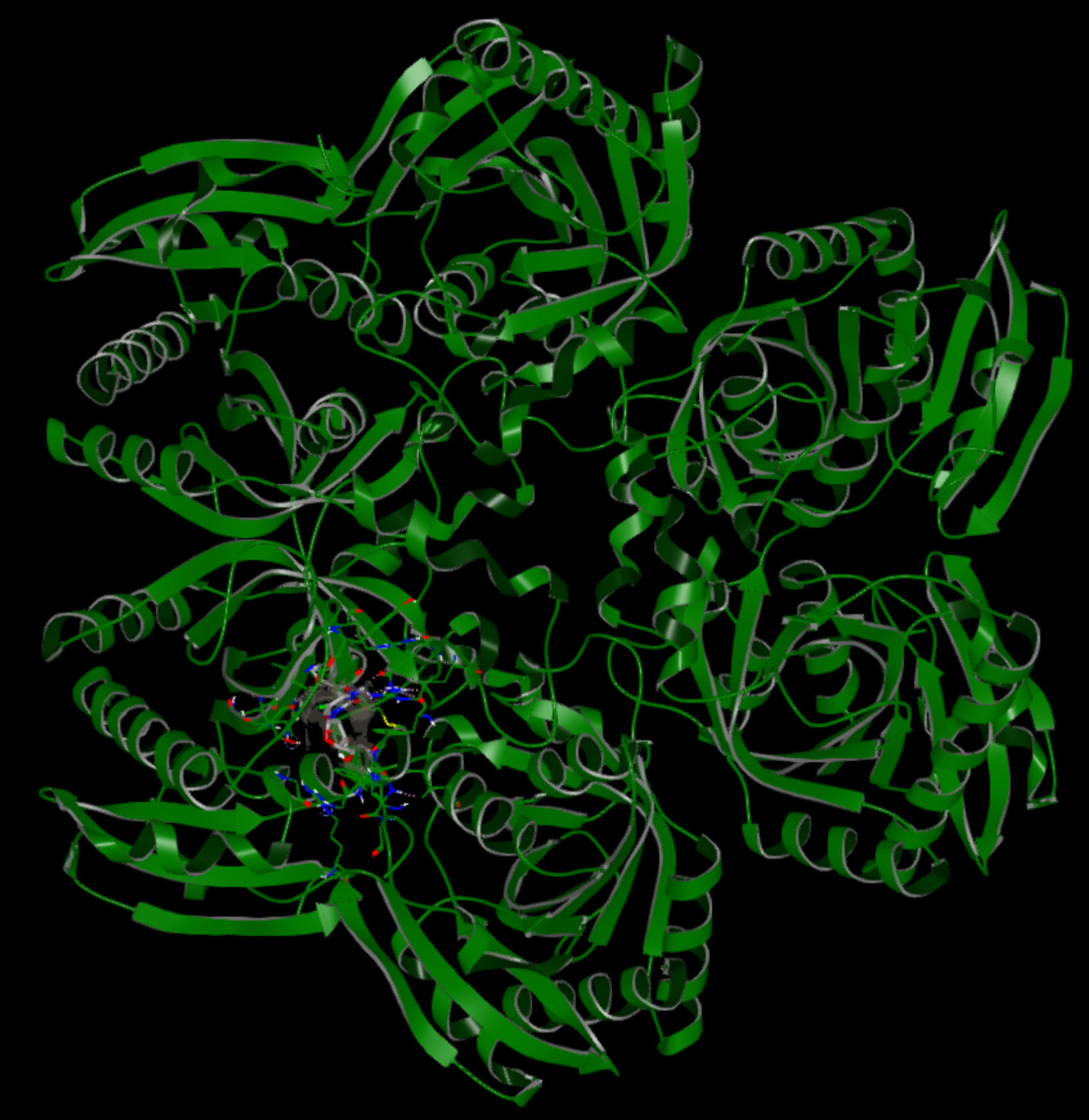
|

|
Structure view |
Binding site view |
SZMAP: Simplified Workflow
SZMAP now provides a simpler but enhanced workflow, using the newly released SPRUCE technology for structure preparation.
Structures prepared in the OEDU file format from SPRUCE can be used as direct input
to SZMAP or GamePlan without the need for any further preparation steps.
The single utility application Spruce4Szmap replaces the three previous utility applications Pch,
MKHetDict, and FixDupAtomNames. Spruce4Szmap also eliminates the need to use
the external program reduce as part of structure preparation workflow.
SZYBKI: SMIRNOFF
The 2019.Nov release incorporates the ability to use force fields based on SMIRNOFF
(SMIRks Native Open Force Field) specifications into SZYBKI and FreeForm.
Two such small molecule force fields, smirnoff99Frosst and OpenFF1.0.0,
from the Open Force Field Initiative have been
incorporated in this release. Both the smirnoff99Frosst and OpenFF1.0.0 force
fields can handle almost all pharmaceutically relevant chemical space.
The SMIRNOFF specification differs from traditional force field specifications principally in the data representation of the chemistry. It uses extended SMARTS strings for parameters instead of atom types, greatly reducing the number of interdependent parameters.
The Open Force Field Initiative is actively working on improvements to OpenFF1.0.0, and
promises to bring newer force fields in the future. OpenEye plans to keep up to date with the latest
versions of these force fields as they become available. A future release will incorporate
the ability to allow using custom versions of SMIRNOFF force fields in SZYBKI.
General Notices
Windows installation behavior has changed. Installation of OpenEye-applications bundle will now replace the previous version. See Installation for more information.
All applications were ported to build using OEToolkits 2019.Oct. The previous versions were built using OEToolkits 2019.Apr.
Platform Support
RHEL6 is no longer supported.
Windows 7 is no longer supported.
RHEL8 is now partially supported (command line applications only). Qt applications will be included in future releases once Qt is supported on RHEL8.