Filtering¶
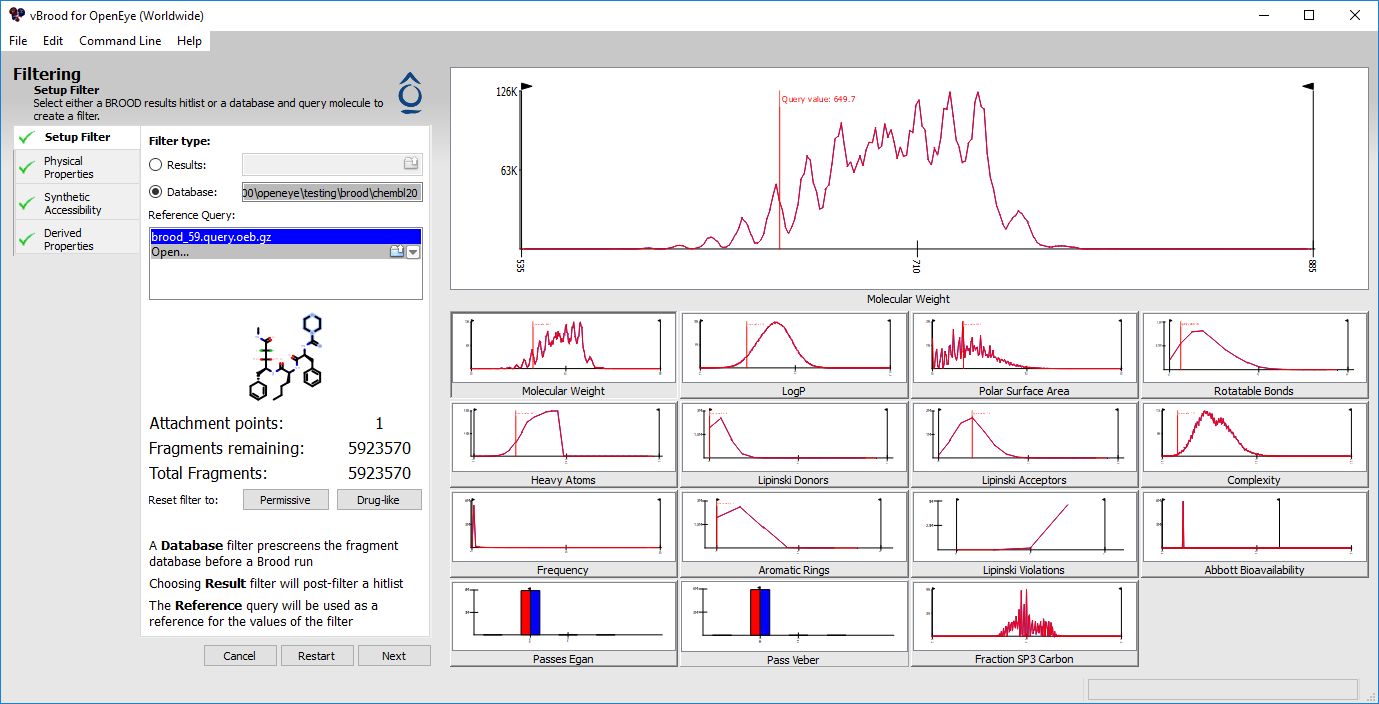
The vBROOD filter interface¶
vBROOD provides tools for creating filters for BROOD runs and for post-filtering hitlists. The filtering tool displays the distributions of the values of various properties both pre- and post- filter. The values can be adjusted numerically or visually. The various properties are available in three separate tabs:
Physical properties
Synthetic accessibility
Derived properties
Building a filter for running BROOD and filtering a hitlist are much the same process, though the initial setup is a bit different (see below).
You can select the desired path by selecting the appropriate radio button: Results for filtering a hitlist or Database for building a BROOD run filter.
For details on specific property filters, see Property Selection.
The Filter window¶
The Filter window consists of 12 histograms detailing various medicinal chemistry properties. While filtering, each histogram displays two lines, one red and one blue. The red line indicates the pre-filter distribution. The blue line indicates the distribution of the remaining fragments after all filters have been applied. Additionally, when creating a BROOD Run filter, the histograms will display a red vertical line at the value for the query molecule.
The large histogram at the top is interactive and is a reproduction of one of the smaller 11 histograms in the bottom half of the screen. To set the property to be displayed in the primary histogram, click on one of the smaller histograms.
The top histogram is interactive and has one or two draggable black lines capped with Left and Right Arrows. Clicking and dragging one of these lines can set the minimum or maximum value for the given property. Changing this value will also update the histograms of all the other properties to give an estimate of the distribution of the values remaining.
Filtering¶
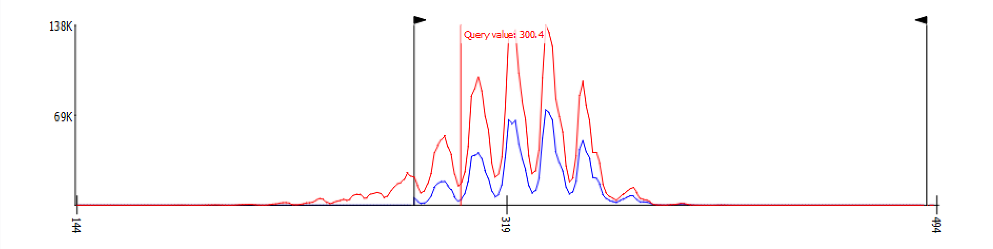
A property histogram¶
Filter values can be set two ways: by typing a value or by dragging the filter line. To set by value, navigate to the page containing the property and adjust the appropriate control to set the value.
There is one Setup Filter page and three property pages. The Setup Filter page allows you to choose the type of filtering you want and to perform filter setup. The three property pages are Physical Properties, Synthetic Accessibility and Derived Properties. Each of these pages provides direct input of filter ranges by typing in exact values.
For details about specific property filters, see Property Selection.
Setting up a filter¶
The first page is the Setup Filter page that allows you to choose whether to build a run filter or to perform a hitlist filtering. It also displays how many fragments or molecules are remaining, and allows the current filter to be set to one of the two presets, Permissive and Druglike.
BROOD Run filter¶
Building a filter allows BROOD to quickly eliminate some fragments of the database without doing the 3D overlap. This can result in much shorter run times. The filter is built in the context of a query, though it may be used with any query. The values in vBROOD are not in the same reference frame as the values passed to BROOD. vBROOD combines query information to allow users to generate filters at the molecular scale rather than the fragment scale. When vBROOD calls BROOD, it translates the filter to fragment-based values by removing the query contribution.
To build a filter for BROOD:
Select the Database radio button.
Select a query. You can select a query from the query list, open a query from a file using the Open item in the query list, or load a recently opened query from the Recents menu.
Once a query has been selected, vBROOD will populate the histograms.
Hitlist filtering¶
Hitlists can also be filtered after a BROOD run:
Select the Results radio button.
Open a BROOD Results file by clicking on the Open folder.
After loading, the histograms will be populated with the values from the Results file. On the final page (Derived Properties), choose Save Hitlist to save your filtered hitlist to disk.
Filter presets¶
The Druglike preset filter sets the property filters to the following ranges:
Synthetic Accessibility => .1
Heavy Atom Count <= 30
Molecular Weight <= 500
Rotor Count <= 10
LogP <= 5.0
Polar Surface Area <= 150
Lipinski Donor Count <= 5
Lipinski Acceptor Count <= 10
Lipinski Failures <= 1
ABS >= .25
Fraction sp3 Carbons >= .3
Aromatic Freres Jacques <= 4
Egan egg is not required
Veber bioavailability is not required
The Permissive filter sets property ranges so all fragments through pass the filter.