OEApplications 2024.2¶
Release Highlights 2024.2.1¶
The OEApplications 2024.2.1 is a bug-fix of the OEApplications 2024.2.0 release, and depends on OEToolkits 2024.2.1.
Release Highlights 2024.2.0¶

OMEGA: Thompson Sampling for Torsion Driving¶
A Bayesian approach to explore conformer space with torsion driving has been added as an alternative
to the exhaustive searching of the space in OMEGA. This uses the framework of Thompson sampling
to quickly direct the conformer search towards the low energy structures, reducing the time required
to find the lowest energy conformer. As such, Thompson sampling is particularly suitable for generating
small to moderately sized ensembles, such as used in the FastROCS or ROCS mode.
Thompson sampling provides significant speedup for such ensemble generation, particularly for molecules
with a large number of rotors.
Conformer ensembles for FastROCS usage are generated with OMEGA using both the standard
exhaustive sampling and the newly implemented Thompson sampling, for ~5500 Iridium HT protein bound
small molecule crystal structures. Performance between the two sampling methods is compared
in terms of RMSD between the generated ensemble and the crystal pose, and in the time required to
generate the ensembles. The results of the comparison, as shown in Figure 1 below, show that
the accuracy of the generated ensembles, measured in RMSD, remains unaffected when using
exhaustive versus Thompson sampling, whereas there is significant reduction in time usage to
generate those ensembles when using Thompson sampling, especially as the size of the molecules grows.
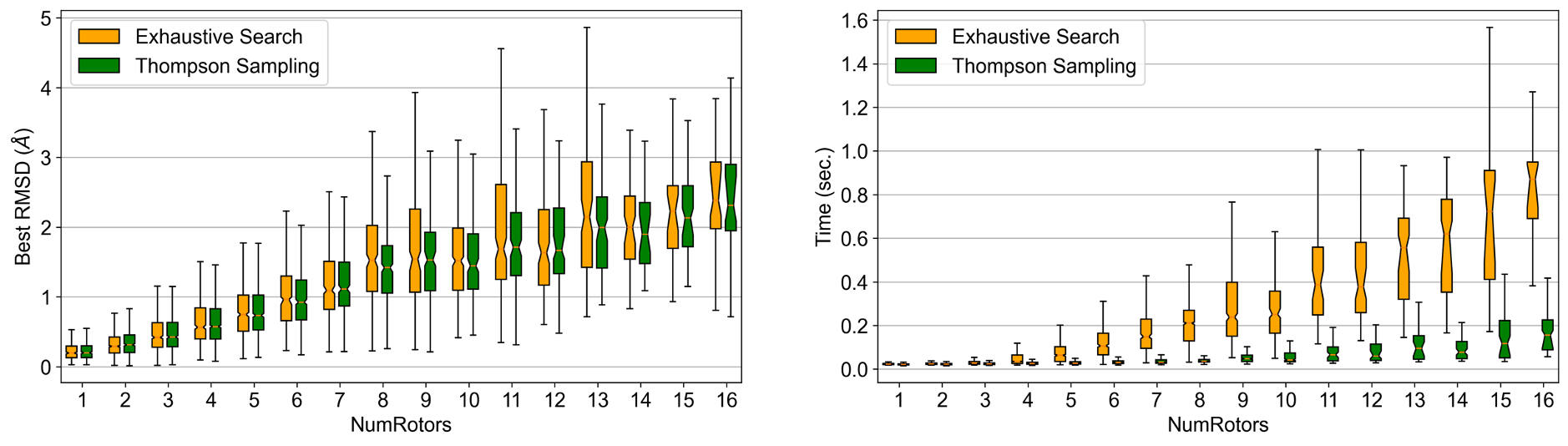
Figure 1. Comparison of OMEGA performance between exhaustive and Thompson sampling.¶
Ligand-based virtual screening performance, using both the exhaustive and Thompson sampling generated ensembles, was also compared and displayed in Figure 2. The comparisons were performed using directory of useful decoys (DUD-Z) datasets containing 38 targets. The results ensure that there is no meaningful change in accuracy when using Thompson sampling for ensemble generation for usage in ligand-based virtual screening.
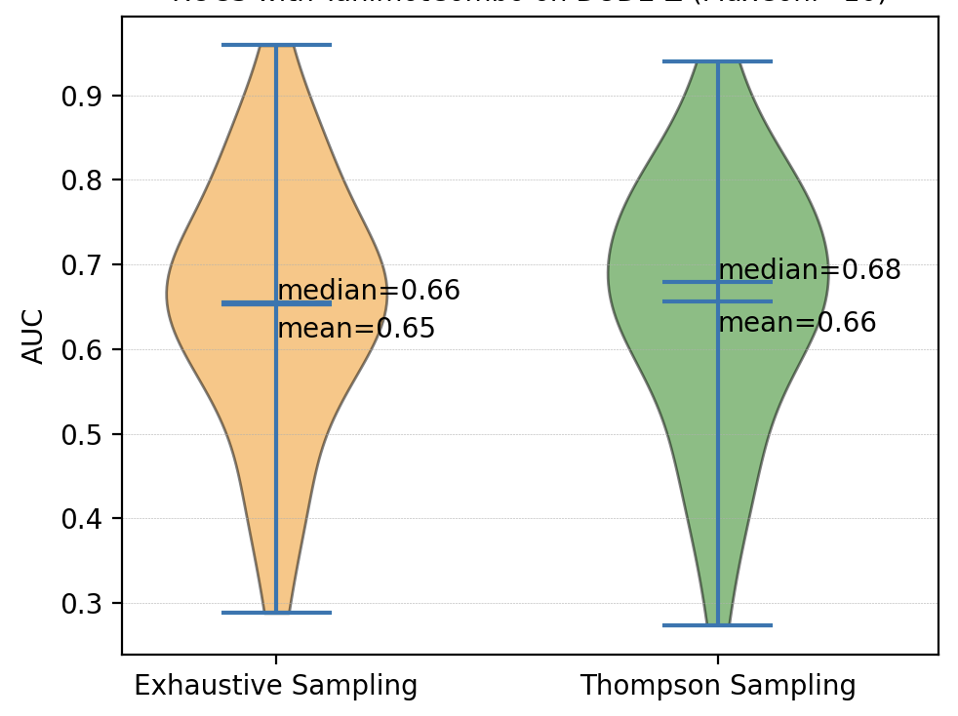
Figure 2. Comparison of performance of ligand-based virtual screening success rate with conformer ensembles generated using exhaustive versus Thompson sampling.¶
BROOD: Bioisosteric Fragment Replacement in Macrocyclic Peptides¶
A new pose generation algorithm has been introduced in Bioisostere TK, as well as in BROOD, based on flexible overlay of shape, color, and force fields, as used in pose generation with ShapeFit. The new algorithm generates high quality poses of molecules that are obtained from fragment replacement in Bioisostere TK and BROOD. The robust pose generation algorithm enables usage of Bioisostere TK and BROOD for molecules with complex geometry, including macrocyclic peptides. Figure 3 shows an example of a residue fragment replacement in a macrocyclic peptide.
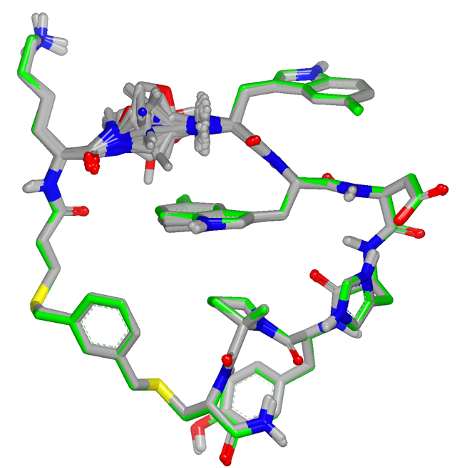
Figure 3. Bioisosteric replacement of a residue fragment in a macrocyclic peptide using BROOD.¶
Eon TK: New Toolkit for Overlay Optimization with Shape and Charge Density¶
The 2024.2 release introduces Eon TK, a new toolkit for molecular similarity based on shape and electrostatics. This provides toolkit-level access to the existing EON application functionality. Following EON, the new toolkit has both molecular charge density and electrostatic potential as options to use as descriptors of electrostatic properties. Figure 4 shows an example of EON overlay with charge density visualization.
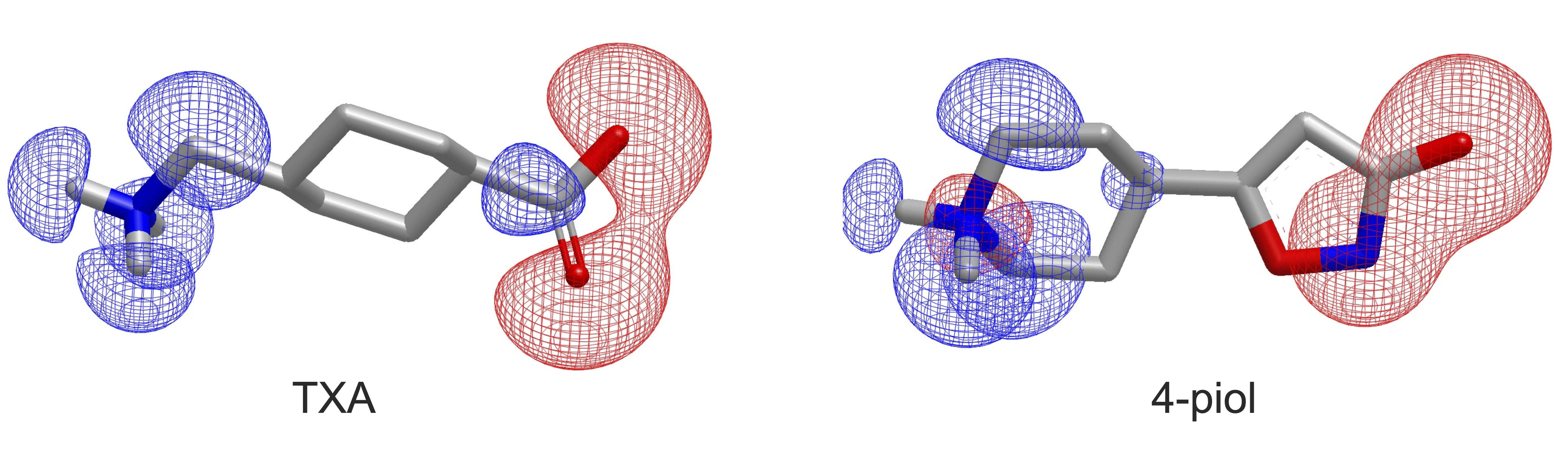
Figure 4. EON shape charge density overlay between two molecules.¶
Eon TK provides tools for molecular overlay optimization with shape and charge density, similarity estimation based on shape and charge density or electrostatic potential, and corresponding hit list building.
Supported Platforms¶
OS
Versions
Linux
RHEL8/9, Ubuntu20/22
Windows
Win10, Win11
macOS
12, 13, 14
Detailed Release Notes 2024.2¶
AFITT 3.0.1¶
Minor internal improvements have been made.
BROOD 4.1.0¶
Minor internal improvements have been made.
Bioisostere TK 4.1.0¶
New features¶
A new algorithm, based on OEFlexiOverlay, has been adapted in Build for optimization of generated poses for molecules obtained from fragment replacement. The new algorithm is more robust for optimization of poses for all geometries, including cyclic peptides.
Major bug fixes¶
Query building algorithms in OECreateBroodQuery have been improved for robustness. The improved API automatically includes any associated hydrogen atoms as part of the query fragment. Behavior has also been improved to provide better error behavior against invalid selections.
The new methods SetSingleSourceMol and GetSingleSourceMol have been added to OEFragmentOptions to provide flexibility regarding source molecule storage during database building.
EON 3.1.0¶
New Features¶
Supported input and output file formats for EON have been revised to improve clarity.
The output csv file now contains additional columns with a rank of hits and best conformer index.
The Boolean flag
-scoreonlyhas been added to the EON application to allow passing prealigned molecules that only need to be scored.
Bug Fixes¶
An issue affecting the EON hit list has been fixed. EON now properly uses the best conformer for each database molecule based on shape and charge density scores, instead of the first conformer of the molecule, to generate the hit list.
A bug in combining shape and electrostatics gradients has been fixed.
Eon TK 3.1.0¶
This is the first commercial release of Eon TK.
New features¶
The following classes are available as preliminary API with the first release of this toolkit.
The following functions are available as preliminary API with the first release of this toolkit.
Zap TK 2.5.0¶
Minor internal improvements have been made.
OEDocking 4.3.2¶
Minor internal improvements have been made.
OEDocking TK 4.3.2¶
New features¶
The method Optimize in OEPoseOptimizer now returns an
unsignedcode corresponding to any possible failure, instead of abool. The method also takes additional arguments for protein and ligand masks in the OEDesignUnit.The following methods have been added to the OESinglePoseResult:
A new method, MethodChoice, has been added to OEPosit that provides information on the method that would be used for the pose prediction of a given ligand against a given receptor.
The new methods GetTorLib and SetTorLib have been added to OEPositOptions to provide a choice for the torsion library that would be used for conformer generation during pose prediction.
The following methods have been added to the OEShapeFitOptions:
Major bug fixes¶
The method Optimize in OEPoseOptimizer is now more robust in optimizing a docked pose. Accordingly, Docking with relaxation in OEPosit is also now more robust.
The function OEHasClash now properly recognizes all clashes between a ligand and a protein that were previously mistaken because of some misidentification of covalent bonds.
OMEGA 6.0.0¶
New Features¶
Supported input and output file formats for OMEGA have been revised to improve clarity.
Minor Bug Fixes¶
OMEGA now properly propagates
SD dataof molecules when running jobs usingMPI.OMEGA no longer reports time used per molecule in the log output, but they are properly reported in the
CSVreport.
Omega TK 6.0.0¶
New features¶
The following new methods have been added to OETorDriveOptions to enable use of Thompson sampling, instead of exhaustive sampling, of torsion angles during torsion driving. Use of Thompson sampling is especially useful, being significantly faster without losing quality, in generating smaller ensembles as obtained with the FastROCS or ROCS modes.
A new class, OEThompsonOptions, has been added that enables controlling Thompson sampling during torsion driving.
Generation of a single conformer with OEConformerBuilder has been modified to use Thompson sampling.
Minor bug fixes¶
User-defined values passed through SetEnergyWindow, SetMaxConfs, and SetRMSThreshold are now properly accounted for, even when the corresponding defaults are range-based.
Molecule titles are retained when Build fails in OEOmega with
fails to build structure from CT.A verbose log is provided for fragments that fail when Build fails in OEOmega with
fails to build structure from CT.The OEOmegaIsGPUReady function has been safeguarded from a rare crash. The function has also been updated to check not only for device 0, but for devices from 0 to 100.
MolProp TK 2.6.5¶
Minor internal improvements have been made.
PICTO 5.1.2¶
New features¶
A feature has been added that allows a user, in the view preferences, to increase the limit on the maximum number of atoms in a molecule when reading or writing in molecule-specific file formats.
OEDepict TK 2.5.5¶
Minor internal improvements have been made.
pKa-Prospector 1.2.4¶
Minor internal improvements have been made.
QUACPAC 2.2.5¶
Minor internal improvements have been made.
Quacpac TK 2.2.5¶
Minor bug fixes¶
Some issues with primary amines processing in OEMultistatepKaModel have been fixed.
ROCS 3.7.0¶
New Features¶
The utility application CheckCff has been restructured to provide the same underlying functionality for color assignment as that of the Prep method from the OEOverlapPrep class. The following new command line parameters have been added.
-removeDups: Flag if color atoms within a certain distance from each other should be removed.-minDistance: Minimum distance between color atoms for them to be treated as duplicates.
The modified CheckCff always generates a report file in
.csvformat and-reportcan be used to provide a nondefault name to the report file.
Shape TK 3.7.0¶
New features¶
A new method, AddColorGaussians, has been added that allows adding color Gaussians to the OEShapeQuery based on intensity of values on a grid.
A new class, OEOverlapPrepOptions, has been added that allows modifying options that would be used during the Prep. Subsequently, a new constructor for OEOverlapPrep has been added that takes the OEOverlapPrepOptions as an argument.
Minor bug fixes¶
Color force field names in OEColorFFParameter are now treated as case-insensitive.
SiteHopper 2.1.1¶
Minor internal improvements have been made.
SiteHopper TK 2.1.1¶
Minor bug fixes¶
The OESiteHopperIsGPUReady function has been safeguarded from a rare crash.
SPRUCE 1.6.1¶
Minor internal improvements have been made.
New features¶
Command line arguments have been added to control the gap and extend penalties for the sequence alignment used for loop building.
Spruce TK 1.6.1.1¶
Major bug fixes¶
An issue has been fixed where OEExtractBioUnits function would fail if there was a mismatch in author assigned chain names and PDB assigned chain names in the case where a molecule was extracted from an mmCIF file.
An issue was fixed in OEProtonateDesignUnit where a rule to flip clashing in-plane hydrogens on double bonded nitrogens was triggering incorrectly in specific instances.
Spruce TK 1.6.1¶
New features¶
OEBuildSidechains now minimizes target side chains by default. Minimization can be controlled by setting minimizeSidechains and minimizeSidechainsShell options in the OESidechainBuilderOptions class.
A feature has been added to support a next generation mmCIF feature, such that when an mmCIF file provides a connectivity record for the heterogens, Spruce will no longer look up the heterogen in the Chemical Component Dictionary but trust the input provided.
A feature has been added that allows in-plane flipping of a single hydrogen on a double bonded nitrogen, when a clash is detected as part of hydrogen bond network optimization.
Major bug fixes¶
An issue has been fixed related to using the OEHeterogenMetadata class, where the provided SMILES string was not respected for tautomer generation; when tautomers were not provided; and when the title (residue name) collided with existing residue names in the stored Chemical Component Dictionary.
An issue has been fixed related to covalently bound ligands, where the pre-reaction input SMILES and related tautomers were not adjusted properly when accounting for the formed covalent bond.
An issue has been fixed where the Spruce filter deleted single (disconnected) amino acid residues when they were marked HETATMs and were not real “floating” residues in part of the regular protein sequence.
Minor bug fixes¶
OEMutateResidues, OEMutateResidue, and OEBuildSidechains now reduce failures arising from clashes by using shell minimization by default.
An issue has been fixed that caused HEME-like molecules to be excluded from the stored chemical component database.
Documentation changes¶
Information has been added about setting and getting minimizeSidechains and minimizeSidechainsShell options in the OESidechainBuilderOptions class.
SZMAP 1.7.1¶
Minor internal improvements have been made.
Szmap TK 1.7.1¶
Minor internal improvements have been made.
SZYBKI 2.8.0¶
Minor internal improvements have been made.
Szybki TK 2.8.0¶
New features¶
The constant SAGE_OPENFF refers to the latest version of the Sage OpenFF force field.
VIDA 5.0.7¶
Minor internal improvements have been made.
Previous Release Highlights¶
- Release Highlights 2024.1.3
- Release Highlights 2024.1.0
- Supported Platforms
- Related Application Versions
- Release Highlights 2023.2.3
- Release Highlights 2023.1.1
- Release Highlights 2023.1.0
- Release Highlights 2022.2.2
- Release Highlights 2022.2.1
- Release Highlights 2022.1
- Release Highlights 2021.2
- Release Highlights 2021.1
- Release Highlights 2020.2.2
- Release Highlights 2020.2
- Release Highlights 2020.1.1
- Release Highlights 2020.1
- Release Highlights 2020.0
- Release Highlights 2019.Nov
- Release Highlights 2019.May
- OEApplications 2018.Nov